Taxonomic Or FUnctional Metagenomic Assembly and PrOfiling = TOFU-MAaPO
This pipelines analyses short reads and can perform analysis of taxonomic profiling, abundance of microbial metabolic pathways and more.
Documentation about the pipeline can be found in the docs/ directory or under the links below:
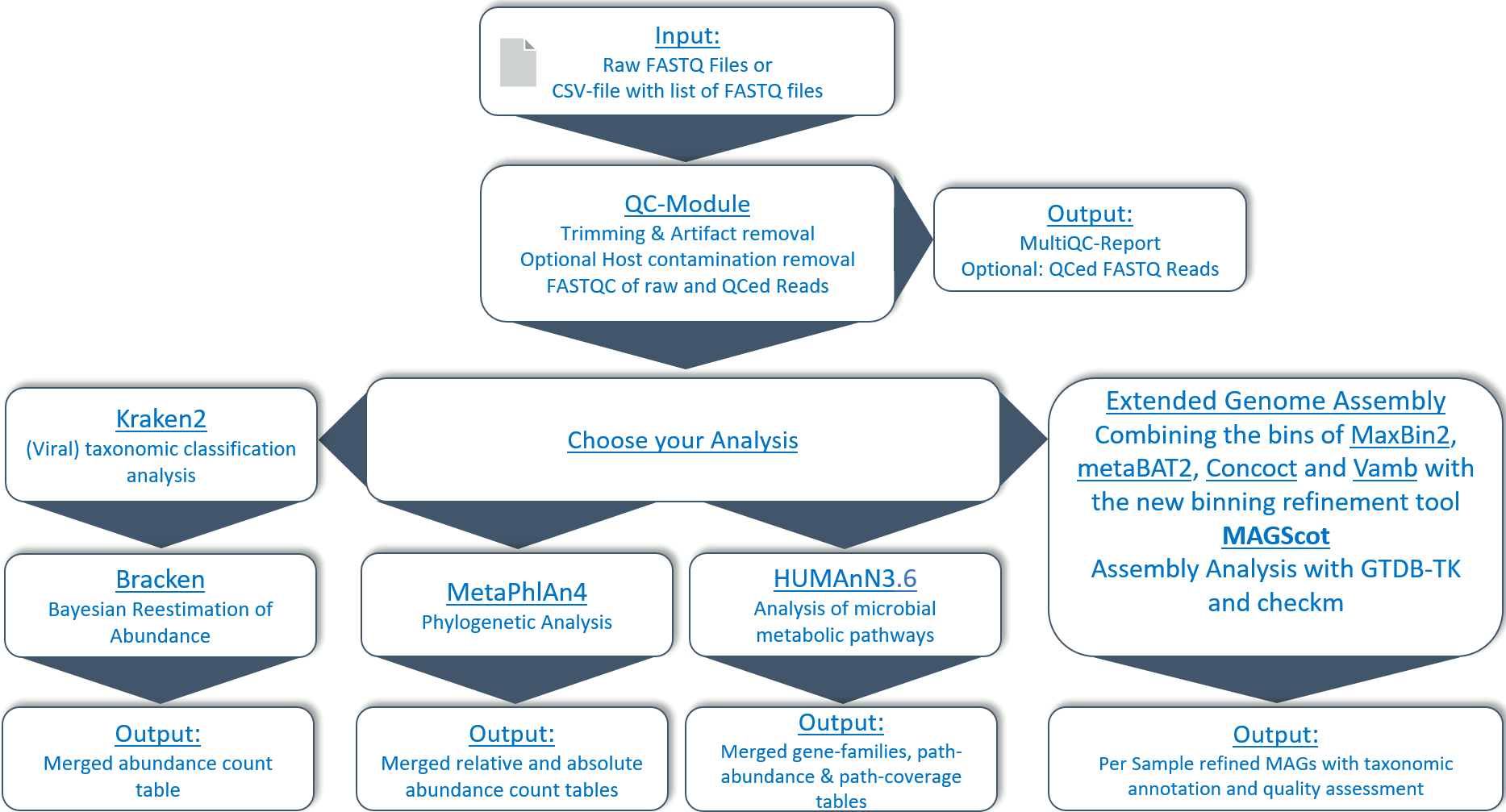
With the help of Docker containers, the pipeline can be run on any common operation system. The user only needs to install Nextflow and Singularity as dependencies and can then run the pipeline without further installation. The pipeline can download all required databases and can remove optionally the host genome reads from the sequencing files and Kraken2 databases for execution. Only one command needs to be run to install, initialise and run the pipeline from start to finish. The pipeline requires short single- or paired-end metagenomic shotgun sequencing FASTQ files as input, or a single csv file containing a list of samples and their associated FASTQ files. The pipeline processes the data for quality control and possible host decontamination (optional) and performs downstream analysis for taxonomic abundance profiles of each sample using Kraken2, Bracken and/or Metaphlan4, metabolic pathway analysis using Humann (v3.6) or two workflows for metagenomic genome assembly. Genome assembly can be performed with a simple approach using Megahit to generate contigs. Contigs are catalogued and indexed using minimap2 and then binned using Metabat software. The bins are taxonomically annotated with GTDB-TK and quality checked with checkm. An extended genome assembly approach can also be activated. In this case, the contigs catalogued and indexed with Megahit and minimap2 are not only processed further with Metabat2, but also with Concoct, Maxbin and vamb. The resulting bins from the individual binning tools are then refined and, where possible, combined with MAGScoT based on sets of single-copy microbial marker genes from the Genome Taxonomy Database. The profile of existing marker genes in each binning result from the different algorithms is compared, and new hybrid candidate bins are created if the binsets have a user-adjustable proportion of marker genes. The results are also taxonomically annotated with GTDB-TK and quality checked with checkm. An estimated bin coverage per sample is generated as additional output.
The project was funded by the German Research Foundation (DFG) Research Unit 5042 - miTarget INF.