Kraken2 classification
A Snakemake pipeline wrapper of the Kraken2 short read metagenomic classification software, with additional tools for analysis, plots, compositional data and differential abundance calculation. Designed and maintained by Ben Siranosian in Ami Bhatt's lab at Stanford University.
Introduction
Kraken2 is a short read classification system that is fast and memory efficient. It assigns a taxonomic identification to each sequencing read, by using the lowest common ancestor (LCA) of matching genomes in the database. Using Bracken provides accurate estimates of proportions of different species. This guide will cover some of the basics, but the full Kraken2 manual has much more detail.
NEW AS OF 2019-09-01!
The outputs of this pipeline have been vastly improved! Both internally and saved data now use the GCTx data format, from the CMapR package. Basically, a GCT object is a data matrix that has associated row and column metadata. This allows for consistent metadata to live with the classification data, for both the rows (taxonomy information) and columns (sample metadata). See section 8. GCTx data processing for more information and tools for working with the new implementation.
Also as of this update, the NCBI taxonomy information used by Kraken is filtered and improved some before saving any data or figures. For example, there were previously many taxonomy levels simply labeled "environmental samples" that are now named with their pared taxa name to remove ambiguity. Also, levels without a proper rank designation (listed with an abbreviation and a number in the kraken report) have been forced into a specific rank when nothing was below them. This makes the taxonomy "technically incorrect", but much more practically useful in these cases. Contact me with any questions. The full list of changes is described in Additional considerations
Table of contents
- Installation
- Usage
- Available databases
- Downstream processing and plotting
- Additional considerations
- Expanded database construction
- Using metagenome assembled genomes as a database
- GCTx data parsing
Quickstart
Install
If you're in the Bhatt lab, most of this work will take place on the SCG cluster. Otherwise, set this up on your own cluster or local machine (with a small database only). You will have to build a database, create the taxonomy_array.tsv file (instructions at previous link), set the database options, and create the sample input files. Thanks to Singularity, all you need to have installed is snakemake. See the instructions here to set up snakemake and set up a profile to submit jobs to the cluster.
Then clone the repo wherever is convenient for you. I use a directory in ~/projects
cd ~/projects
git clone https://github.com/bhattlab/kraken2_classification.git
Run
Copy the config.yaml file into the working directory for your samples. Change the options to suit your projects and make sure you specify the right samples.tsv file. See Usage for more detail. You can then lauch the workflow with a snakemake command like so:
# Snakemake workflow - change options in config.yaml first
snakemake -s path/to/Snakefile --configfile config.yaml --use-singularity --singularity-args '--bind /oak/,/labs/,/home' --profile scg --jobs 99
After running the workflow and you're satisfied the results, run the cleanup command to remove temporary files that are not needed anymore.
snakemake cleanup -s path/to/Snakefile --configfile config.yaml
Parsing output reports
The Kraken reports classification/sample.krak.report, bracken reports sample.krak_bracken.report, and data matrices or GCTx objects in the processed_results folder are the best for downstream analysis. See Downstream processing and plotting for details on using the data in R.
Example output
The pipeline outputs data, results and figures in the structure below.
| classification
- sample.krak Kraken results - classification of each read. These files
can get very large and are unnecessary if you only want the reports.
- sample.krak.report Kraken report - reads and percentages at each taxonomic level.
- sample.krak.report.bracken Standard bracken report at species level. Not useful, use the one below.
- sample.krak_bracken.report Most useful format of the the Bracken results.
| processed_results
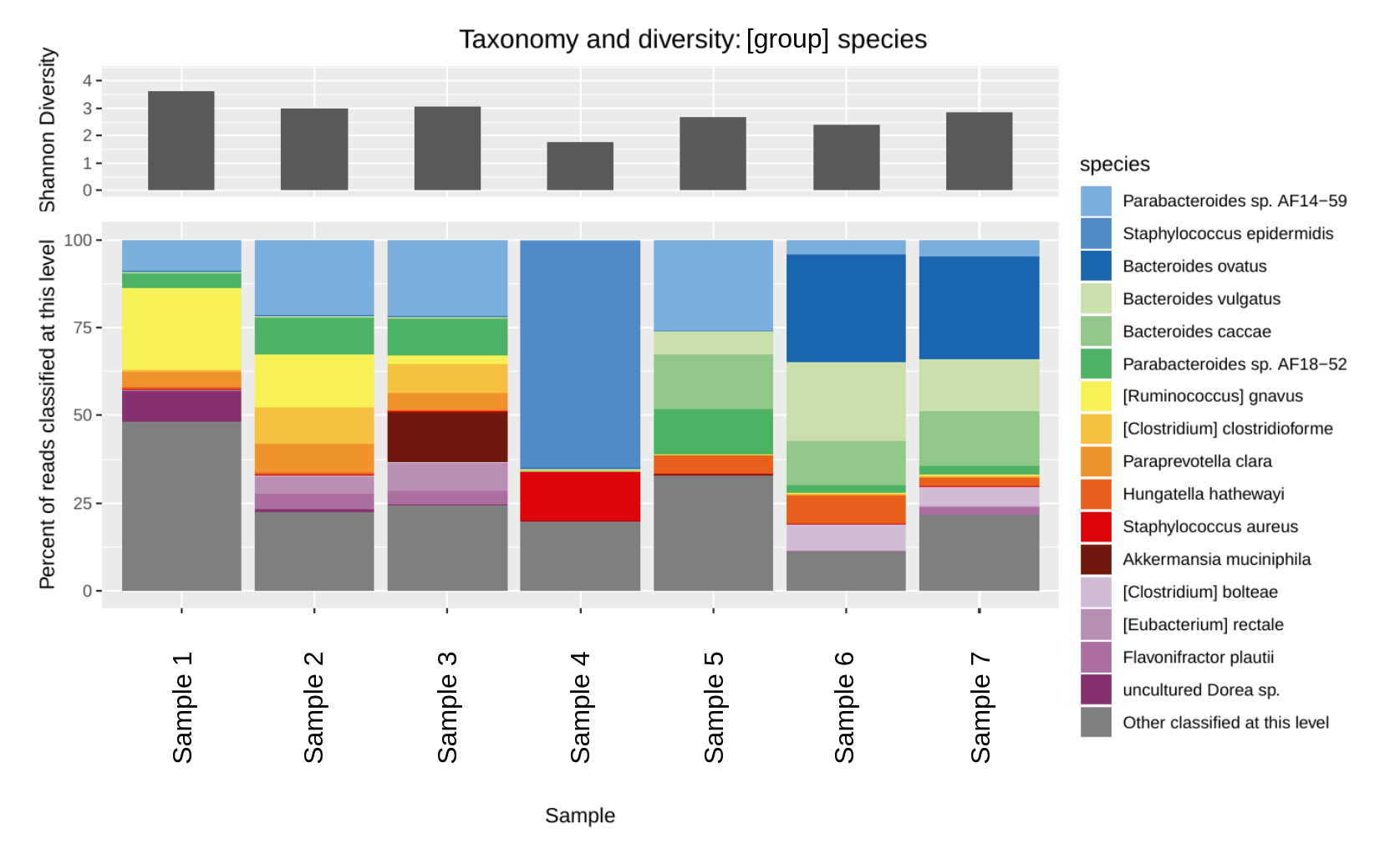
- diversity.txt Diversity of each sample at each taxonomic level
| ALDEX2_differential_abuncance Compositional data analysis done with the ALDEx2 package.
Only done if you have 2 groups in the sample_groups file.
- aldex_result_[].tsv Differential abundance at the given taxonomic level.
- aldex_scatter_[].pdf Scatterplot of effect vs dispersion with significant hits highlighted
- aldex_significant_boxplots_[].pdf Boxplot of any significant hits
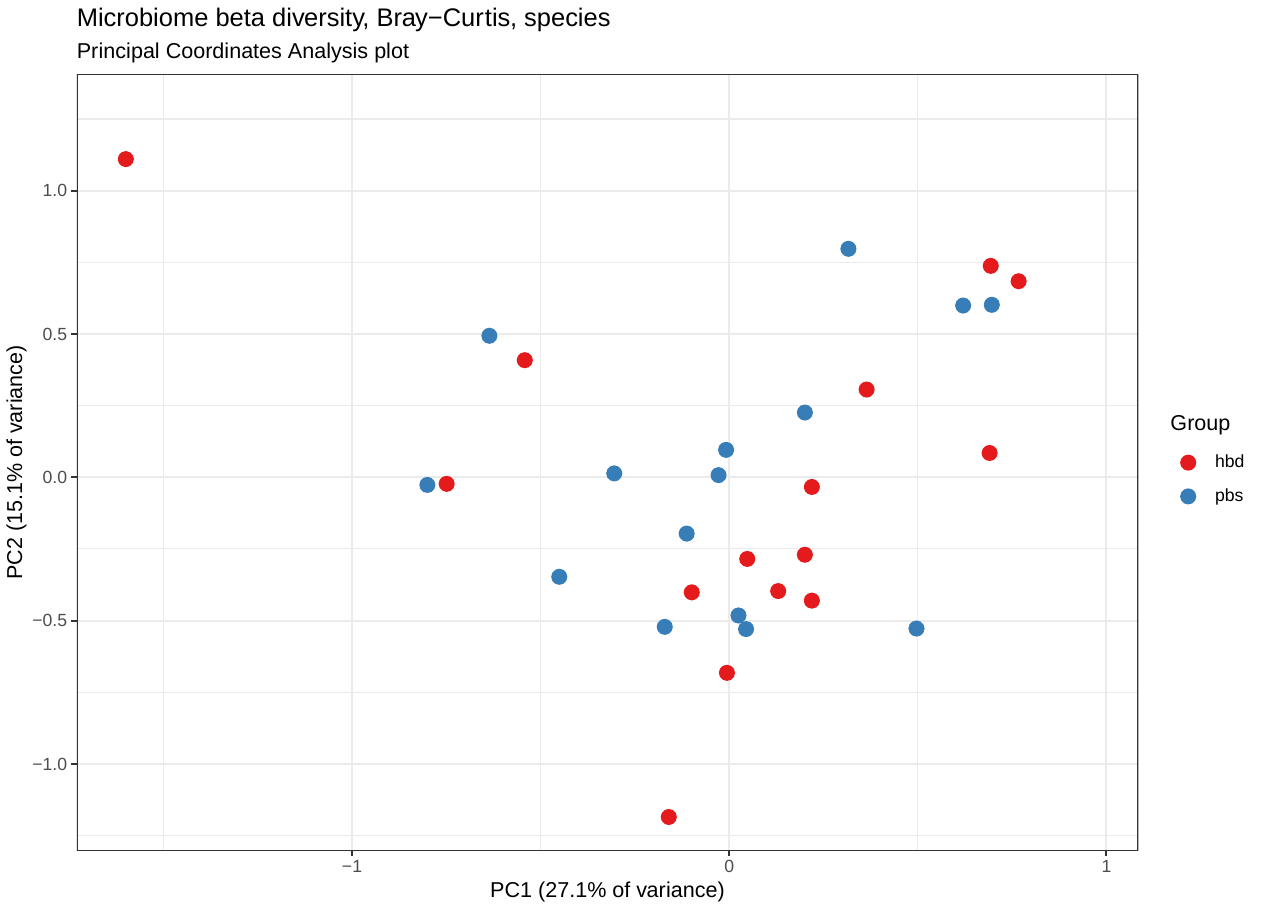
| braycurtis_matrices
- bravcurtis_distance_[].tsv Matrix of braycurtis distance between samples at each taxonomic level
| plots Lots of plots!
- classified_taxonomy_barplot_[].pdf Barplot at each taxonomic level.
- compositional_PCA_plot.pdf PCA done on clr values
- diversity_allsamples.pdf Diversity barplot
- diversity_by_group.pdf Diversity barplot, stratified by sample group
- PCoA_2D_plot_labels.pdf Principal coordinates analysis, calculated on braycurtis distances
- PCoA_2D_plot_nolabels.pdf Same as above without the point labels
- rarefaction_curve.pdf Rarefaction "collectors" curve plot
| taxonomy_gctx_classified_only
- bracken_[]_reads.gctx GCTx file with matrix containing reads classified at each level
- bracken_[]_percentage.gctx Same, but percentage of classified reads
| taxonomy_matrices_classified_only
- bracken_[]_reads.txt Matrix of taxa by sample classified reads
- bracken_[]_percentage.txt Same, but percentage of classified reads
- clr_values_[].txt From compositional data analysis, centered log-ratio values at each level
| processed_results_krakenonly
| Same as above, but using the results without Bracken. Also includes taxonomy matrices
| that have unclassified reads in them (as bracken no longer reports unclassified reads)
| unmapped reads
- sample_unmapped_1.fq Only present if selected in the config file; reads that are not
- sample_unmapped_2.fq classified, as paired end fastq.