ICLR 2024
Authors: Shengchao Liu+, Jiongxiao Wang+, Yijin Yang, Chengpeng Wang, Ling Liu, Hongyu Guo*, Chaowei Xiao*
+ Equal contribution
* Equal advising
[Paper] [Project Page] [ArXiv]
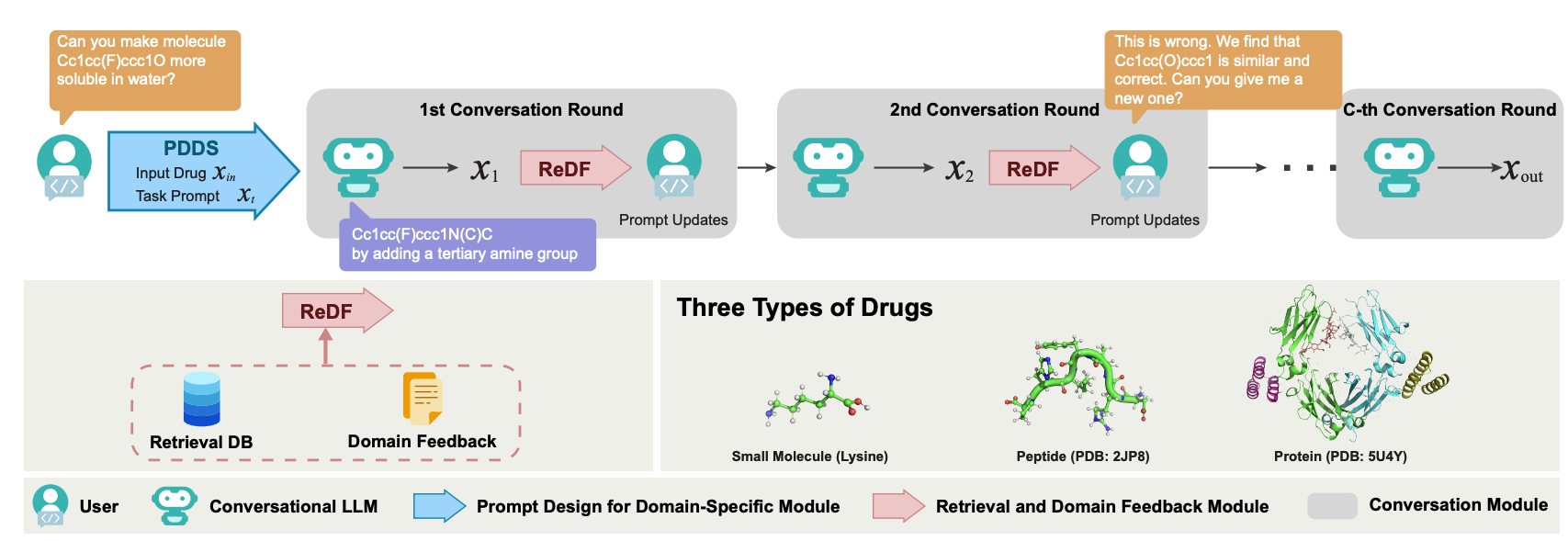
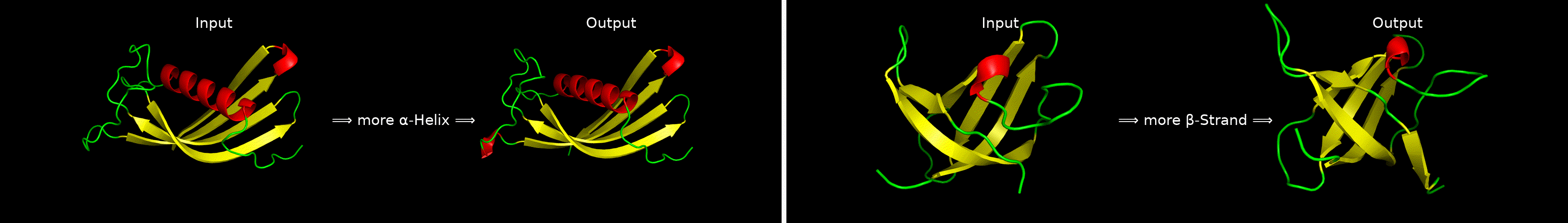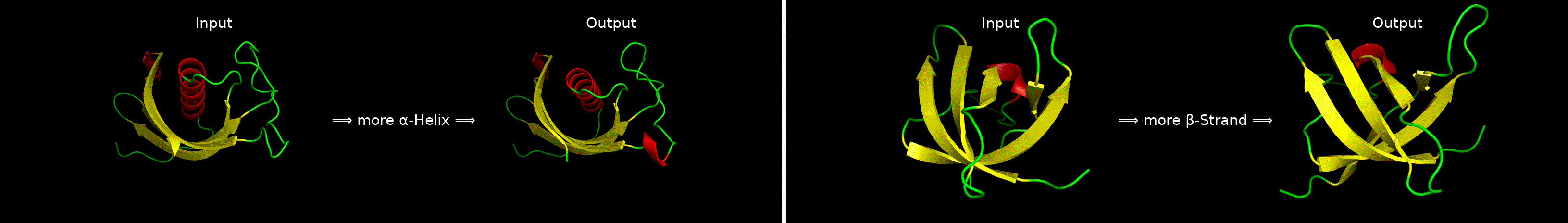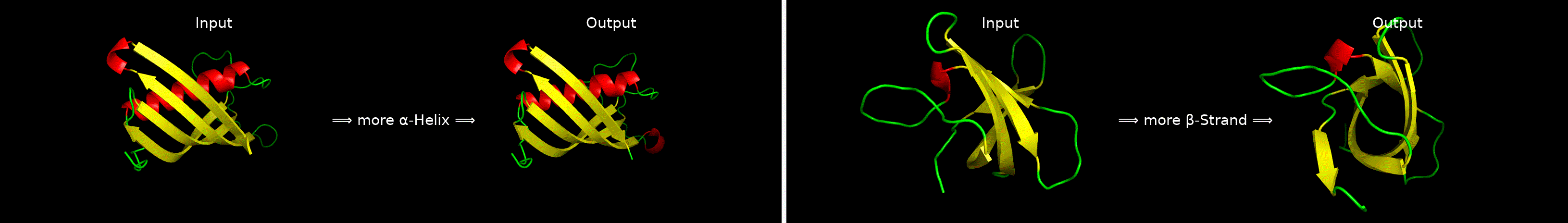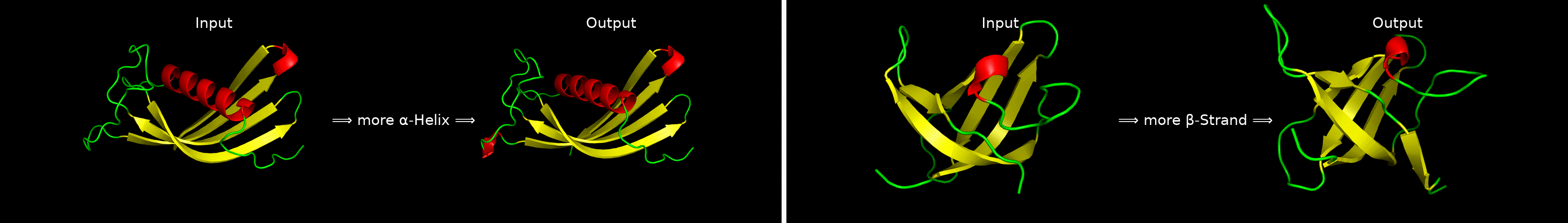
ChatDrug is for conversational drug editing, and three types of drugs are considered:
- Small Molecules
- Peptides
- Proteins
Setup the anaconda (skip this if you already have conda)
wget https://repo.continuum.io/archive/Anaconda3-2019.10-Linux-x86_64.sh
bash Anaconda3-2019.10-Linux-x86_64.sh -b
export PATH=$PWD/anaconda3/bin:$PATHThen download the required python packages:
conda create -n ChatDrug python=3.8
conda activate ChatDrug
pip install rdkit-pypi==2022.9.4
conda install -y numpy networkx scikit-learn
conda install -y -c conda-forge -c pytorch pytorch=1.9.1
pip install tensorflow
pip install mhcflurry
pip install levenshtein
pip install transformers
pip install lmdb
pip install seqeval
pip install openai
pip install fastchat
pip install psutil
pip install accelerate
pip install -e .We provide the dataset in this link. You can manually download and move to the data folder or using the following python script.
from huggingface_hub import snapshot_download
snapshot_download(repo_id="chao1224/ChatDrug_data", repo_type="dataset", local_dir="data", local_dir_use_symlinks=False, ignore_patterns=["README.md"])
Please give credits to the original papers. For more details of dataset, please check the data folder.
The evaluation metrics for three editing tasks are below:
| Drug Type | Evaluation |
|---|---|
| Small Molecule | RDKit (conda install -y -c rdkit rdkit) |
| Peptide | MHCFlurry |
| Protein | ProteinDT paper, checkpoints |
For evaluation on peptides and proteins, please read the following instructions:
- For peptides (MHCFlurry), please run the following bash commands:
> pip install mhcflurry
> mhcflurry-downloads fetch models_class1_presentation
> mhcflurry-downloads path models_class1_presentation
$PATH
> mv $PATH data/peptide/models_class1_presentation
- For proteins (ProteinDT / ProteinCLAP), please run the following python script:
from huggingface_hub import hf_hub_download
hf_hub_download(
repo_id="chao1224/ProteinCLAP_pretrain_EBM_NCE_downstream_property_prediction",
repo_type="model",
filename="pytorch_model_ss3.bin",
cache_dir="data/protein")
Please give credits to the original papers. For more details of evaluation, please check the data folder.
All the task prompts are defined in ChatDrug/task_and_evaluation. you can also find it on the hugging face link.
Please provide your OpenAI API Key in ChatDrug/task_and_evaluation/Conversational_LLMs_utils.py
To use ChatDrug, please use the following command:
python main_ChatDrug.py --task task_id --log_file results/ChatDrug.log --record_file results/ChatDrug.json --C 2
Results will be saved in results/.
For protein editing tasks, multiple evaluation times in retrieval process would consume a lot of time. Thus, we provide a fast version of conversation setting. Running the following command to implement accelerate ChatDrug for protein editing tasks:
python main_ChatDrug.py --task task_id --log_file results/ChatDrug_fast_protein.log --record_file results/ChatDrug_fast_protein.json --C 2 --fast_protein
We also provide code for In-Context Learning setting:
python main_InContext.py --task task_id --log_file results/InContext.log --record_file results/InContext.json
Feel free to cite this work if you find it useful to you!
@inproceedings{liu2024chatdrug,
title={Conversational Drug Editing Using Retrieval and Domain Feedback},
author={Shengchao Liu, Jiongxiao Wang, Yijin Yang, Chengpeng Wang, Ling Liu, Hongyu Guo, Chaowei Xiao},
booktitle={The Twelfth International Conference on Learning Representations},
year={2024},
url={https://openreview.net/forum?id=yRrPfKyJQ2}
}