AISTATS 2022
Authors: Shengchao Liu, Meng Qu, Zuobai Zhang, Huiyu Cai, Jian Tang
[Project Page] [Paper] [ArXiv] [Code] [NeurIPS AI4Science Workshop 2021]
This repository provides the source code for the AISTATS'22 paper Structured Multi-task Learning for Molecular Property Prediction, with the following contributions:
- To our best knowledge, we are the first to propose a new research problem: multi-task learning with an explicit task relation graph;
- We construct a domain-specific multi-task dataset with relation graph for drug discovery;
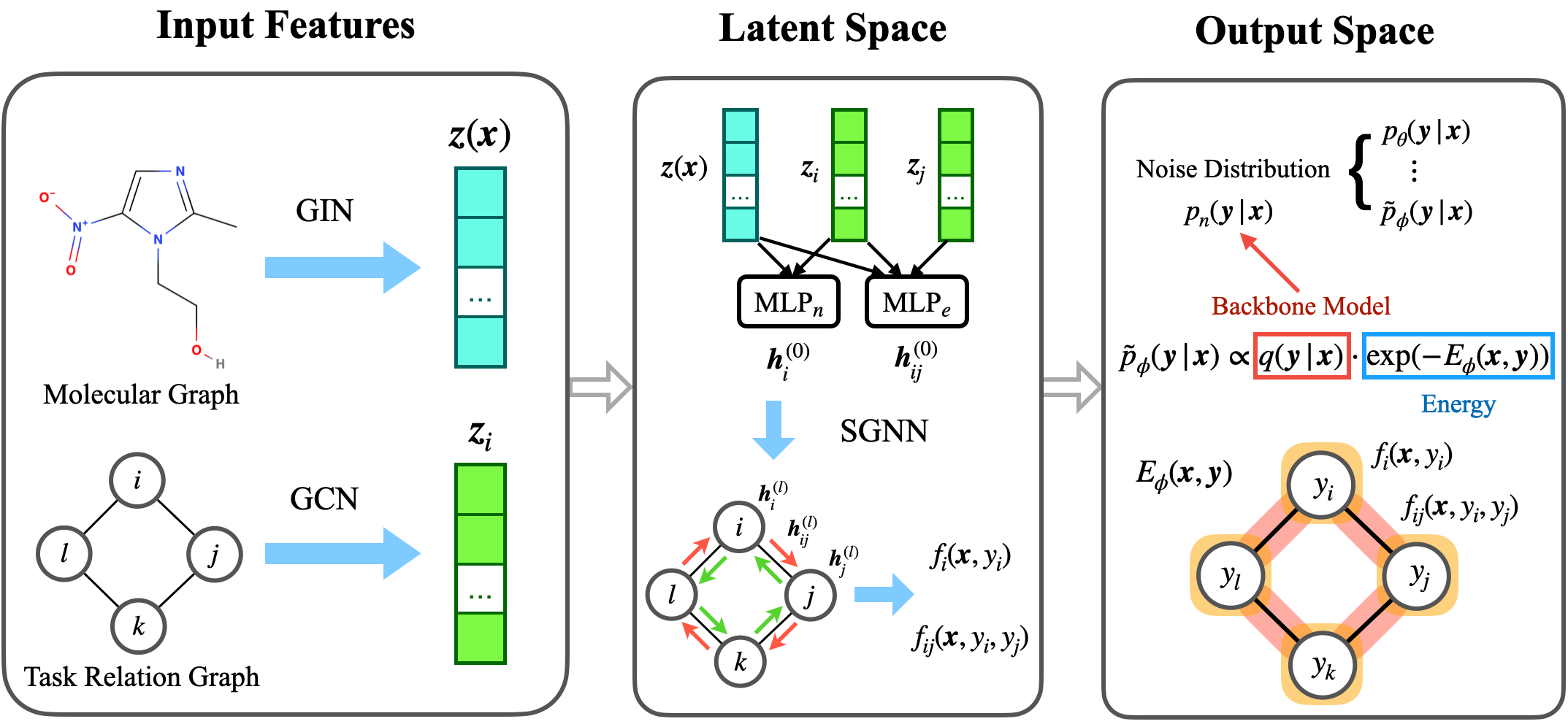
- We propose state graph neural network-energy based model (SGNN-EBM) for task structured modeling in both the latent and output space.
In the future, we will merge it into the TorchDrug package.
For implementation, this repository also provides the following multi-task learning baselines:
- standard single-task learning (STL)
- standard multi-task learning (MTL)
- Uncertainty Weighing (UW)
- GradNorm
- Dynamic Weight Average (DWA)
- Loss-Balanced Task Weighting (LBTW)
Below is environment built with pytorch-geometric.
conda create -n SGNN_EBM python=3.7
conda activate SGNN_EBM
conda install -y -c pytorch pytorch=1.6.0 torchvision
conda install -y matplotlib
conda install -y scikit-learn
conda install -y -c rdkit rdkit=2019.03.1.0
conda install -y -c anaconda beautifulsoup4
conda install -y -c anaconda lxml
wget https://data.pyg.org/whl/torch-1.6.0%2Bcu102/torch_sparse-0.6.9-cp37-cp37m-linux_x86_64.whl
wget https://data.pyg.org/whl/torch-1.6.0%2Bcu102/torch_scatter-2.0.6-cp37-cp37m-linux_x86_64.whl
wget https://data.pyg.org/whl/torch-1.6.0%2Bcu102/torch_spline_conv-1.2.1-cp37-cp37m-linux_x86_64.whl
wget https://data.pyg.org/whl/torch-1.6.0%2Bcu102/torch_cluster-1.5.9-cp37-cp37m-linux_x86_64.whl
pip install torch_sparse-0.6.9-cp37-cp37m-linux_x86_64.whl
pip install torch_scatter-2.0.6-cp37-cp37m-linux_x86_64.whl
pip install torch_spline_conv-1.2.1-cp37-cp37m-linux_x86_64.whl
pip install torch_cluster-1.5.9-cp37-cp37m-linux_x86_64.whl
pip install torch-geometric==1.6.*
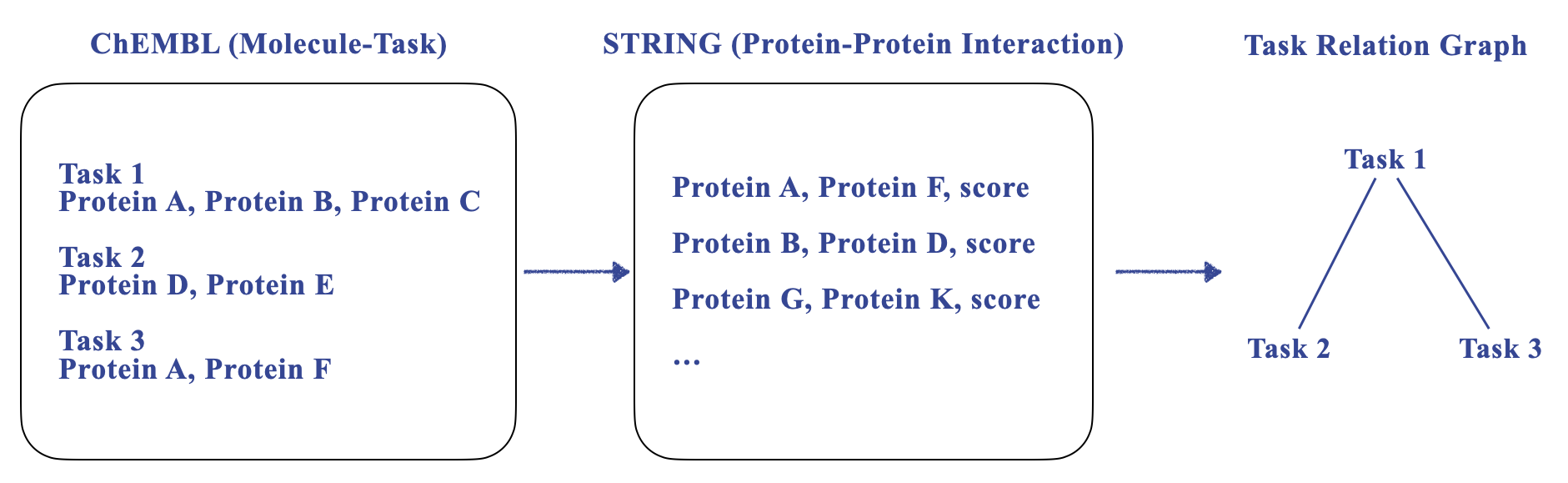
In this work, we propose a novel dataset with explicit task relation. Basically it is a molecule property task dataset, where the task refers to a binary classification problem on a ChEMBL assay. Each task measures certain biological effects of molecules, e.g., toxicity, inhibition or activation of proteins or whole cellular processes, etc. We focus on tasks that target at proteins. Then we extract the task relation by aggregating the protein-protein interaction (PPI, like String dataset) accordingly.
For the detailed pre-processing steps, please check this instruction. The pre-processed datasets can be downloaded here.
We also provide the pre-trained model weights and evaluation scripts accordingly. First you can download the checkpoints here. All the optimal hyper-parameters are provided in the bash scripts.
cd checkpoint
bash eval_SGNN.sh > eval_SGNN.out
bash eval_SGNN_EBM.sh > eval_SGNN_EBM.outHere we provide the script for training the SGNN and SGNN-EBM (adaptive with pre-trained SGNN). Note that the pre-trained SGNN models is required (either using last script or from the pre-trained weights).
bash submit_SGNN.sh
bash submit_SGNN_EBM.shWe also provide the scripts for all six STL and MTL baselines under the scripts folder.
Feel free to cite this work if you find it useful to you!
@inproceedings{liu2022multi,
title={Structured Multi-task Learning for Molecular Property Prediction},
author={Liu, Shengchao and Qu, Meng and Zhang, Zuobai and Cai, Huiyu and Tang, Jian},
booktitle={AISTATS},
year={2022}
}