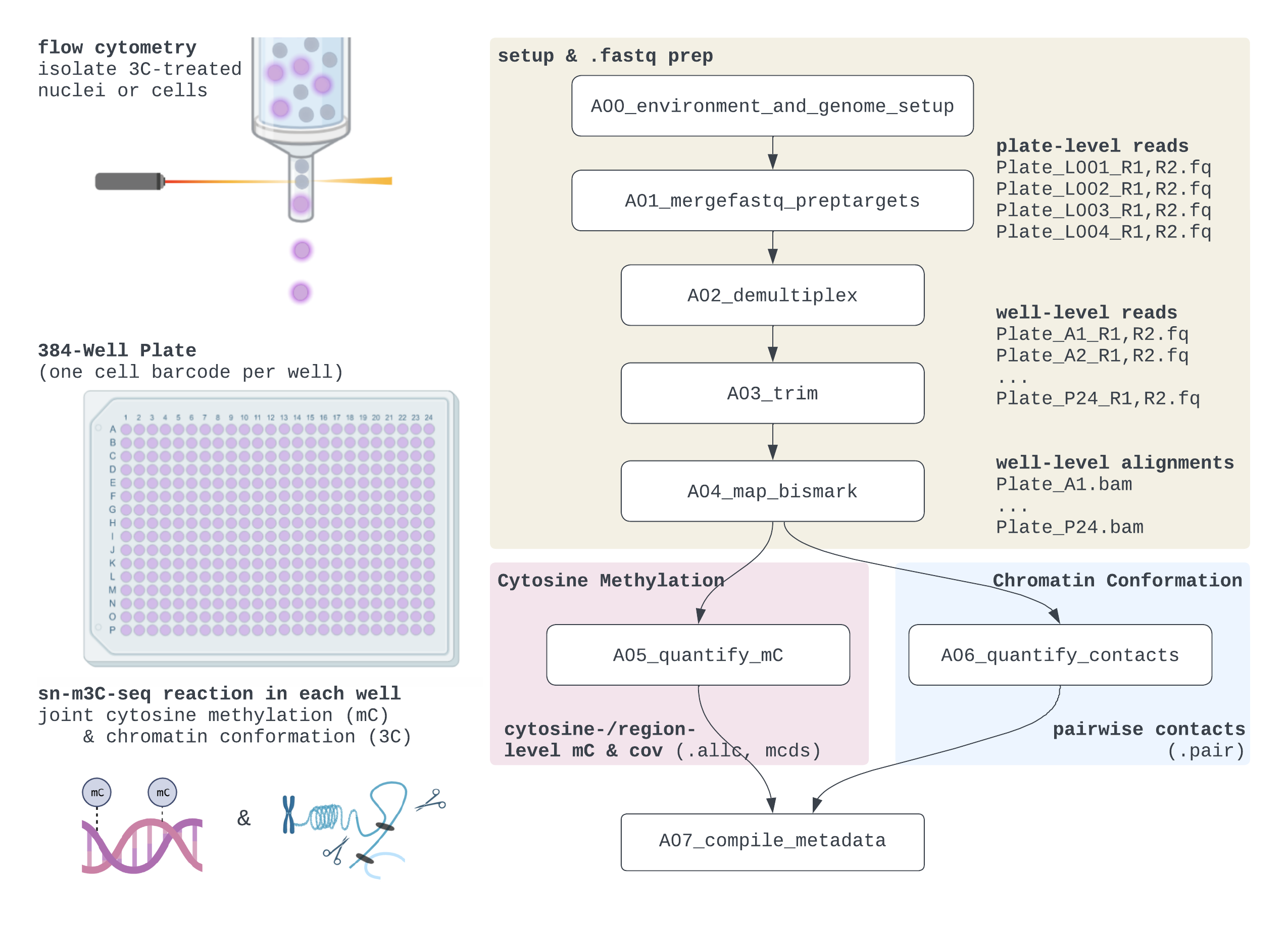
sn-m3C-seq simultaneously profiles DNA methylome (mC) and chromatin contacts within a single nucleus. As DNA methylation is unaltered during chromatin conformation capture (3C) protocols, we perform 3C/HiC-style in situ reactions followed by nucleus isolation into 384-well plates, bisulfite conversion, and sequencing library generation on the 3C ligation products.
What's in this repo?
- An updated version of TAURUS-MH to go from plate-level .fastqs → cell-level alignments (.bam), quality control metrics, and core mC & 3C features.
- For the methylome, the primary features are the methylation counts and coverage for each cytosine in the genome (.allc file), which can then be aggregated into methylation levels of different genomic intervals (e.g., 100kb-bins, genes; .mcds file).
- For chromatin conformation, the features are read-level contacts (similar to .pairs), which can be aggregated into counts in different pairs of bins (10kb, 25kb, 100kb aggregated counts in sparse contact matrices a la .cool files).
What isn't in this repo?
- Cell-level QC steps, which can be celltype- and study-specific. (Some suggestions on the Detailed Overview page and within past manuscripts.)
- Downstream analysis of mC and 3C following basic quantification (e.g., feature selection/calling, clustering, hypothesis testing, imputation).
Quick Start
- Clone (
git clone) or download this repo as a .zip file from the releases page. - install dependencies in .yaml:
conda env create -f Documentation/snm3Cseq_taurus.yml - customize the genome alignment and input sample metadata (either using Jupyter notebooks
A00*andA01*OR by editing ScriptsA00*andA01*) - run submission scripts (
.sub) in order, paying attention to parallelization task IDs-t 1-Nplatesand-t 1-Nbatches(24 wells/batch by default). For convenience, this can be done via the Notebooks, or the full list of submission commands is listed at Documentation/submission_helper.txt. - read the Detailed Overview for FAQs and common pitfalls.
-
Existing quantification pipelines, mainly from our collaborators at/formerly at the Salk Institute:
- TAURUS-MH: read-splitting based mapping pipeline historically used on sn-m3C-seq → developed specifically for this assay primarily by Dr. Dongsung Lee.
- YAP (Yet Another Pipeline): supports sn-m3C-seq and additional related assays (e.g., mC, mCT, mCAT-seq), including new protocol based on restriction-site splitting for m3C. Snakemake. Developed primarily by the Ecker Lab/Drs. Hanqing Liu & Jingtian Zhou.
-
Downstream analysis tools:
- allcools: Typically used by our group for mC downstream analysis. Helpful to review for .allc and .mcds descriptions.
- scHiCluster: Contact pairs to binned contact count matrices at various resolution, including imputation. Our groups have also worked with the Ma Lab's Higashi & FastHigashi.
-
Flagship assay paper and closely related methylome-only reaction:
- sn-m3C-seq: Lee DS, et al. Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nature methods. 2019 Oct;16(10):999-1006.
- snmC-seq2: Luo, C. et al. Robust single-cell DNA methylome profiling with snmC-seq2. Nat. Commun. 9, 7–12 (2018). [Note: we are now on snmC-seq3 but only a wet lab protocol citation exists.]
-
Our library structure is described in the Detailed Overview and a seqspec.
-
Additional examples of applications/publications using sn-m3C-seq:
- Liu, et al. (2023) preprint: Liu H, et al. Single-cell DNA Methylome and 3D Multi-omic Atlas of the Adult Mouse Brain. bioRxiv. 2023 Apr 18.
- Heffel, et al. (2022) preprint: Heffel MG, et al. Epigenomic and chromosomal architectural reconfiguration in developing human frontal cortex and hippocampus. bioRxiv. 2022:2022-10.
- The example in scripts / detailed overview is for a fibroblast → induced pluripotent stem cell (iPSC) differentiation experiment; the underlying data to soon be published for the IGVF Consortium (NHGRI).
- Some raw and processed datasets are linked/publicly available via our lab website luogenomics.github.io. For example, note the
_allc.tar.gz(cytosine-level) and_contacts.tar.gz(contact pairs) files for GEO GSM6596812.
- Thanks to original TAURUS-MH workflow developers, including Drs. Dongsung Lee, Chongyuan Luo, and Jesse Dixon.
- Luo Lab collaborators/members for pipeline testing and feedback, namely Zach von Behren and Min Jen Tsai.