5'-O~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~AAA-3'
O~~~AA O~~ O~ O~~~~~~~AO~~~~~~~~A
O~~ O~~ O~~ O~~ O~O~~ O~~ O~~
O~~ O~~ O~ O~~ O~~ O~~
O~~ O~~ O~~ O~~ O~~~~~AA O~~~~~~A
O~~ O~~ O~~~~~A O~~ O~~ O~~
O~~ O~~ O~~ O~~ O~~ O~~ O~~ O~~
O~~~~A O~~~ O~~ O~~O~~ O~~~~~~~AA
┌─ᐅ 5'-O~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~-3'
...===┴========================================================================================...SCAFE (Single Cell Analysis of Five-prime Ends) provides an end-to-end solution for processing of single cell 5’end RNA-seq data. It takes a read alignment file (*.bam) from single-cell RNA-5’end-sequencing (e.g. 10xGenomics Chromimum®), precisely maps the cDNA 5'ends (i.e. transcription start sites, TSS), filters for the artefacts and identifies genuine TSS clusters using logistic regression. Based on the TSS clusters, it defines transcribed cis-regulatory elements (tCRE) and annotated them to gene models. It then counts the UMI in tCRE in single cells and returns a tCRE UMI/cellbarcode matrix ready for downstream analyses, e.g. cell-type clustering, linking promoters to enhancers by co-activity etc.
- Publications
- Versions
- What does SCAFE do?
- How does SCAFE do it?
- Dependencies
- Installation
- Getting started with demo data
- Run SCAFE on your own data
- Wishlist
Jonathan Moody and Tsukasa Kouno et al. Profiling of transcribed cis-regulatory elements in single cells. bioRxiv 2021.04.04.438388
v1.0.0 (Lastest Stable release) [June 6, 2022]
- auto detects mode for TSO identification and trimming (in scafe.tool.sc.bam_to_ctss)
- use tabix/bgzip for fast indexing of ctss bed
- annotate hyperactive distal locus, analogous to super-enhancer (in scafe.tool.cm.annotate)
- annotate potential alternative promoters beyond reference transcript 5'ends (in scafe.tool.cm.annotate)
- aggregate multiple ctss without considering UMI/Cellbarcode, for scalability, replacing 'pool' tools and workflows (in scafe.tool.cm.aggregate and scafe.workflow.cm.aggregate)
- scafe.tool.sc.link is now obsolete and thus monocle3 and cicero are not needed.
- calculate the directionality of tCREs (in scafe.tool.cm.directionality)
v0.9.0-beta [March 20, 2021]
- Initial pre-release
- beta phase, likely buggy and has performance issues.
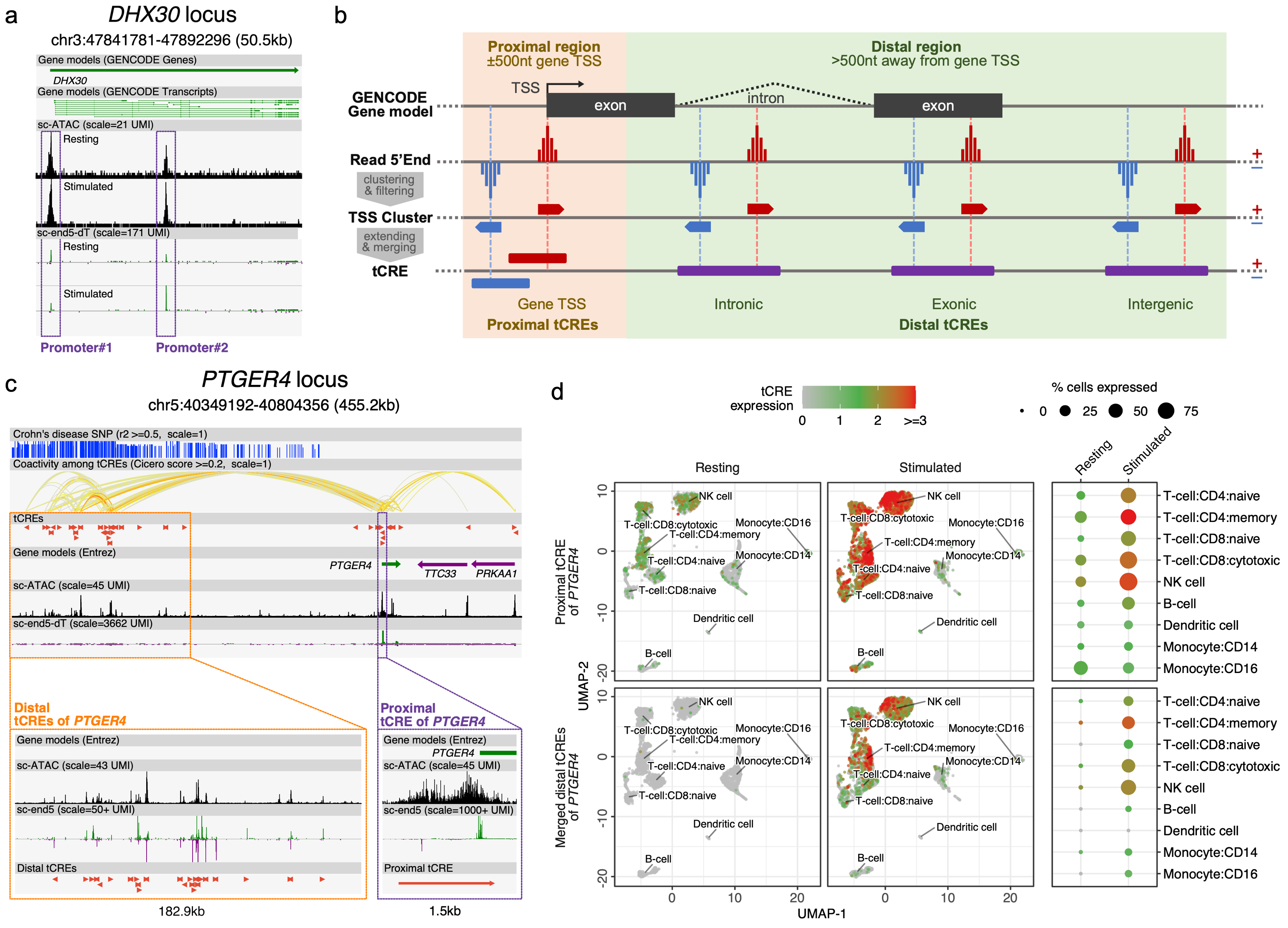
Profiling of cis-regulatory elements (CREs, mostly promoters and enhancers) in single cells allows us to interrogate the cell-type specific contexts of gene regulation and genetic predisposition to diseases. Single-cell RNA-5’end-sequencing methods (sc-end5-seq, available from 10xGenomics Chromimum®) theorectically captures the 5'end of cDNA, which represents transcription start sites (TSS). Measuring the RNA output at TSS allows us to precisely locate transcribed CREs (tCREs) on the genome, enabling the quantification of promoter and enhancer activities in single cells. Figure (a) shows the sc-end5-seq signal at the two promoters of gene DHX30. It highlights the consistency between sc-end5-seq and sc-ATAC-seq data, as well as the dynamic alternaitve TSS usage between cell states (i.e. resting and stimulated immune cells). SCAFE identify genuine TSS information from sc-end5-seq data. Figure (b) illustrates the stretagies of SCAFE to defined tCRE from TSS information. Figure (c) shows the proximal and distal tCREs defined by SCAFE at PTGER4 locus. Figure (d) shows the activities of the proximal and distal tCREs at PTGER4 locus in resting and simulated immune cells, demonstarting the capability of SCAFE to study CRE activities in single cells.
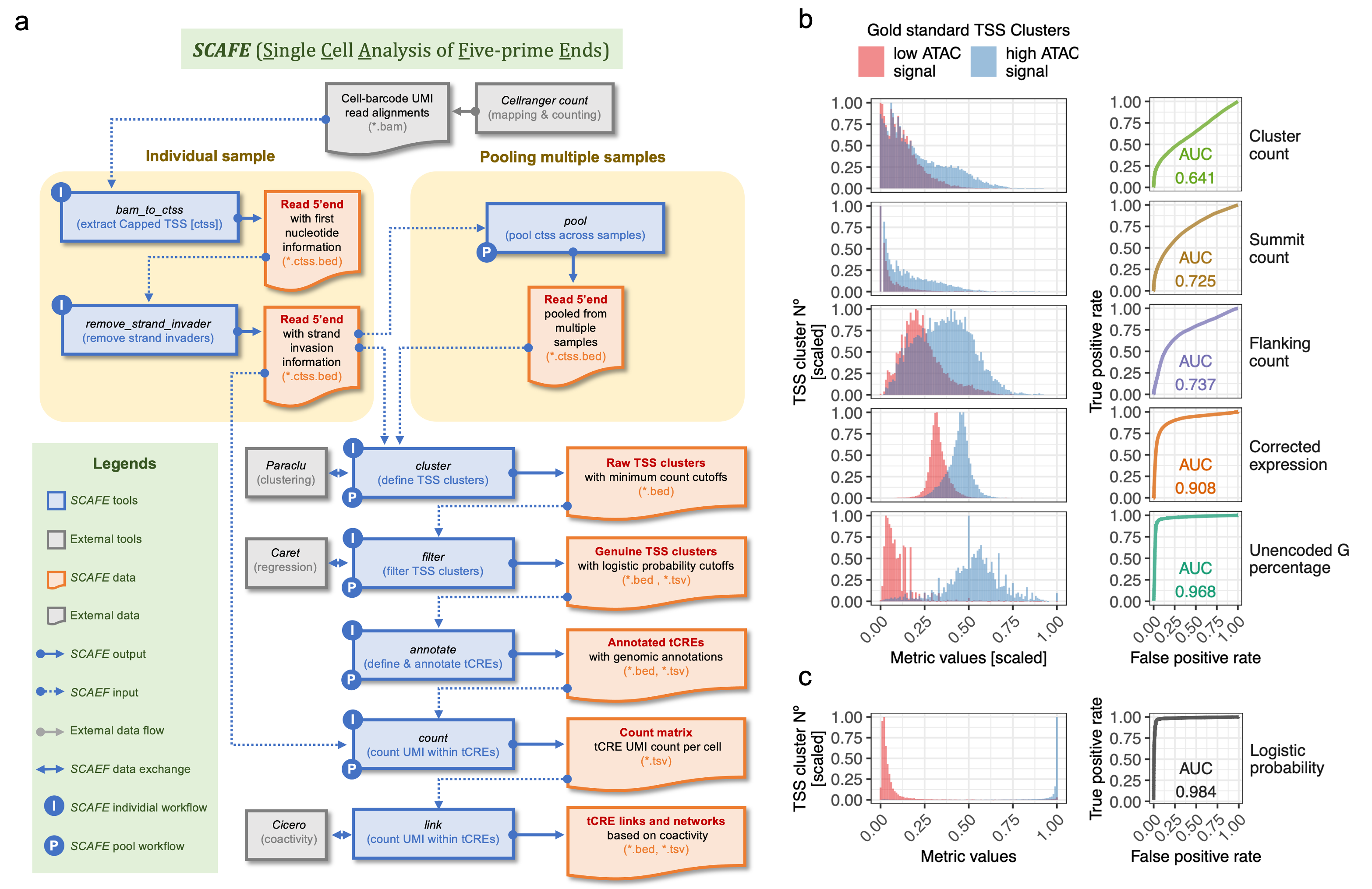
SCAFE consists of a set of perl programs for processing of sc-end5-seq data. Major tools are listed in Figure (a). SCAFE accepts read alignment in *.bam format from 10xGenomics Chromimum® software cellranger. Tool bam_to_ctss extracts the 5’ position of reads, taking the 5’ unencoded-Gs into account. Tool remove_strand_invader removes read 5’ends that are strand invasion artifacts by aligning the TS oligo sequence to the immediate upstream sequence of the read 5’end. Tool cluster performs clustering of read 5’ends using 3rd-party tool Paraclu. Tool filter extracts the properties of TSS clusters and performs multiple logistic regression to distinguish genuine TSS clusters from artifacts. Tool annotate define tCREs by merging closely located TSS clusters and annotate tCREs based on their proximity to known genes. Tool count counts the number of UMI within each tCRE in single cells and generates a tCRE-Cell UMI count matrix. SCAFE tools were also implemented workflows for processing of individual samples or aggregateing of multiple samples.
P.S. SCAFE also accepts bulk 5'end RNA-Seq data (e.g. bulk CAGE). See scripts for details.
A fraction of TSS identified based on read 5′ends from template switching (TS) reactions (used in 10xGenomics Chromimum®) may not be genuine, attributed to various artefacts including strand invasion and other sources. This results in excessive artifactual TSS misidentified along the gene body, collectively known as “exon painting”. While strand invasion artefacts can be specifically minimized by considering the complementarity of TSS upstream sequence to TS oligo sequence, a non-negligible fraction of artefactual TSS remains after filtering for strand invasion. To minimize the artifactual TSS, SCAFE examines the properties of TSS clusters, as shown in Figure (b), and devised a classifier to identify genuine TSS based on multiple logistic regression. This classifier, i.e. logistic probability, achieved excellent performance with AUC>0.98 across sequencing depths and outperformed all individual metrics. This is implemented in the tool filter.
Go straight to the Docker Image section if you do not want to deal with dependencies and already have docker installed.
SCAFE is mainly written in perl (v5.24.1 or later). All scripts are standalone applications and DO NOT require installations of extra perl modules. Check whether perl is properly installed on your system.
#--- Check your perl version
perl --versionSCAFE relies on R for logistic regression, ROC analysis and graph plotting. Rscript (v3.6.1 or later) and the following R packages have to be properly installed:
#--- Check your Rscript version, must be 3.6.1 ot later
Rscript --version
#--- Check your R packages, install if missing
Rscript -e 'install.packages("ROCR")'
Rscript -e 'install.packages("PRROC")'
Rscript -e 'install.packages("caret")'
Rscript -e 'install.packages("e1071")'
Rscript -e 'install.packages("ggplot2")'
Rscript -e 'install.packages("scales")'
Rscript -e 'install.packages("reshape2")'
Rscript -e 'install.packages("R.utils")'Note: scafe.tool.sc.link is now obsolete since v1.0.0 now and thus monocle3 and cicero are not needed.
SCAFE also relies on a number of 3rd party applications. The binaries and executables (Linux) of these applications are distributed with this reprository in the directory ./resources/bin and DO NOT require installations.
SCAFE was developed and tested on Debian GNU/Linux 9, with R (3.6.1) and perl (5.24.1) installed. Running SACFE on other OS with other version of R and perl are not guranteed. In you want to run SCAFE on other OS, we would recommend running it from docker container, see below. If you would like to run SCAFE natively on other OS, you have to ensure the R and perl versions, and might consider downloading and compiling the other 3rd party applications from their own sources. The binaries of 3rd party applications have to be execuatble in SCAFE directoty at ./resources/bin/.
Once you ensured the above dependencies are met, you are ready to download SCAFE to your system.
#--- make a directory to install SCAFE
mkdir -pm 755 /my/path/to/install/
cd /my/path/to/install/
#--- Obtain SCAFE from github
git clone https://github.com/chung-lab/SCAFE
cd SCAFE
#--- export SCAFE scripts dir to PATH for system-wide call of SCAFE commands
echo "export PATH=\$PATH:$(pwd)/scripts" >>~/.bashrc
source ~/.bashrc
#--- making sure the scripts and binaries are executable
chmod 755 -R ./scripts/
chmod 755 -R ./resources/bin/To ensure the dependencies, please run check.dependencies.
#--- run check.dependencies to check
scafe.check.dependenciesIf you have docker installed on your system, you might also consider pulling the SCAFE docker image and run it in a docker container. Once you are logged into the SCAFE docker container, the following tutorial on the demo data can be ran with exactly the same command.
To install docker, please see here. Noted that all files reads/writes are within the docker container by default. To share files (i.e. input and output of SCAFE) between the container and the host, please see here. If you are running Docker on a labtop, make sure the allocated resources (e.g. memory and disk space) are enough, see here. We suggest to allocate at least 16GB of memory.
#---to pull the docker image
docker pull cchon/scafe:latest
#---to run scafe within a docker container, run
docker run -it cchon/scafe:latestThe Docker image can also be used with Singularity container platform, with additional options specified as the followings.
#---to pull the docker image using singularity
singularity pull docker://cchon/scafe:latest
#---to run scafe within a singularity container, run
singularity shell --writable --env APPEND_PATH=/SCAFE/scripts --env LC_ALL=C docker://cchon/scafe:latest
Now you have enssured all dependencies and downloaded SCAFE, time to get the demo data and test a few runs on the demo data.
Demo data and reference genome must be downloaded for testing SCAFE on your system.
#--- download the demo data using script download.demo.input
scafe.download.demo.input
#--- download the reference genome hg19.gencode_v32lift37 for testing demo data
scafe.download.resources.genome --genome=hg19.gencode_v32lift37Now, let's test SCAFE with a workflow (workflow.sc.solo) that processes one library of single cell 5'end RNA-seq data. First we check out the help message of workflow.sc.solo. Remember you can always check the help message for all scripts of SCAFE using the --help flag.
#--- check out the help message of workflow.sc.solo
scafe.workflow.sc.solo --helpIt should print the help message as the followings:
Usage:
5'-O~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~AAA-3'
O~~~AA O~~ O~ O~~~~~~~AO~~~~~~~~A
O~~ O~~ O~~ O~~ O~O~~ O~~ O~~
O~~ O~~ O~ O~~ O~~ O~~
O~~ O~~ O~~ O~~ O~~~~~AA O~~~~~~A
O~~ O~~ O~~~~~A O~~ O~~ O~~
O~~ O~~ O~~ O~~ O~~ O~~ O~~ O~~
O~~~~A O~~~ O~~ O~~O~~ O~~~~~~~AA
┌─ᐅ 5'-O~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~-3'
...===┴========================================================================================...
Single Cell Analysis of Five-prime End (SCAFE) Tool Suite
---> scafe.workflow.sc.solo <---
<--- workflow, single-cell mode, process a single sample --->
Description:
This workflow process a single sample, from a cellranger bam file to tCRE UMI/cellbarcode count matrix
Usage:
scafe.workflow.sc.solo [options] --run_bam_path --run_cellbarcode_path --genome --run_tag --run_outDir
--run_bam_path <required> [string] bam file from cellranger, can be read 1 only or pair-end
--run_cellbarcode_path <required> [string] tsv file contains a list of cell barcodes,
barcodes.tsv.gz from cellranger
--genome <required> [string] name of genome reference, e.g. hg19.gencode_v32lift37
--run_tag <required> [string] prefix for the output files
--run_outDir <required> [string] directory for the output files
--training_signal_path (optional) [string] quantitative signal (e.g. ATAC -logP, in bigwig format), or binary genomic
regions (e.g. annotated CRE, in bed format) used for training of logical
regression model If null, $usr_glm_model_path must be supplied for
pre-built logical regression model. It overrides usr_glm_model_path
(default=null)
--testing_signal_path (optional) [string] quantitative signal (e.g. ATAC -logP, in bigwig format), or binary genomic
regions (e.g. annotated CRE, in bed format) used for testing the performance
of the logical regression model. If null, annotated TSS from $genome will be
used as binary genomic regions. (default=null)
--detect_TS_oligo (optional) [match|trim|skip|auto] in bam_to_ctss step, the modes of detecting TS oligo. 1. match: search for
TS oligo sequence on the read, identify the TSO/cDNA junction as 5'end of
the read. This works only when the error rate of the TS oligo region on
the read is low, otherwise a considerable number of read will be invalid.
2. trim: assuming the 1st N bases of the reads are TS oligo, without
checking the actual sequence. N is determined by the length of TS oligo.
3. skip: assuming the TS oligo was not sequenced, the 1st base of the read
will be treated as the 1st base after the TS oligo. 4. auto: automatically
determines the best mode, best of the observed error rate of the TS oligo
and the frequency of 5'end softclipped bases by the aligner. If softcliped
bases is close to the length of TS oligo, mode 1 or 2 will be chosen,
depending on the observed error rate of the TS oligo (error rate <= 0.1,
mode 1 will be chosen or mode 2 otherwise). If softcliped base os close to
zero, mode 3 will be chosen. (default=auto).
--max_thread (optional) [integer] maximum number of parallel threads, capped at 10 to
avoid memory overflow (default=5)
--overwrite (optional) [yes/no] erase run_outDir before running (default=no)
Dependencies:
R packages: 'ROCR','PRROC', 'caret', 'e1071', 'ggplot2', 'scales', 'reshape2'
bigWigAverageOverBed
bedGraphToBigWig
bedtools
samtools
paraclu
paraclu-cut.sh
tabix
bgzip
For demo, cd to SCAFE dir and run,
scafe.workflow.sc.solo \
--overwrite=yes \
--run_bam_path=./demo/input/sc.solo/demo.cellranger.bam \
--run_cellbarcode_path=./demo/input/sc.solo/demo.barcodes.tsv.gz \
--genome=hg19.gencode_v32lift37 \
--run_tag=demo \
--run_outDir=./demo/output/sc.solo/The help message details the input options, noted some are <required> and (optional). workflow.sc.solo takes a bam file (--run_bam_path=), a cellbarcode list file (--run_cellbarcode_path=), the corresponding reference genome (--genome=) and the output prefix (--run_tag=) and directory (--run_outDir=). At the end of the message, it also prints the commands for running the script on demo data. Now, copy the command and run as the following:
#--- run the workflow on the demo.cellranger.bam, it'll take a couple of minutes
scafe.workflow.sc.solo \
--overwrite=yes \
--run_bam_path=./demo/input/sc.solo/demo.cellranger.bam \
--run_cellbarcode_path=./demo/input/sc.solo/demo.barcodes.tsv.gz \
--genome=hg19.gencode_v32lift37 \
--run_tag=demo \
--run_outDir=./demo/output/sc.solo/If it finishes smoothly, you should see the following message on screen:
#=====================================================
Results of all tasks found. All tasks run successfully
#=====================================================You can also check out some of the outputs:
#--- check the output of tCRE annotation tables
ls -alh ./demo/output/sc.solo/annotate/demo/log
#--- check the output of tCRE UMI count matrix
ls -alh ./demo/output/sc.solo/count/demo/matrixWe recommended a test run for all 5 available workflows on the demo data. It will take around ~20 minutes on a regular system running at 5 threads (default).
#--- check out the help message of demo.test.run
scafe.demo.test.run --help
#--- run the all six available workflows on the demo bulk and single
#--- it'll take a around 20 minutes
scafe.demo.test.run \
--overwrite=yes \
--run_outDir=./demo/output/SCAFE maps the cDNA 5'end by identifying the junction between the TS oligo and the cDNA on Read 1 of sc-end5-seq data on the 10xGenomics Chromimum® platform. Therefore, Read 1 must be sequenced long enough (e.g. >50nt) to allow mappnig to genome. The *.bam files are commonly generated from 10xGenomics Chromimum® cellranger count pipeline. When running cellranger count, the --chemistry option must be SC5P-PE (Pair-end). The *.bam generated from both --chemistry fiveprime and --chemistry SC5P-R2 options are NOT COMPATIBLE with SCAFE as the former will remove the junction between the TS oligo and the cDNA on Read 1 and the latter does not even contain Read 1. If users sequecnced Read 1 only, they could run cellranger count with --chemistry SC5P-PE option by supplying cellranger a dummy Read 2 fastq with Read 1 reverse complemented. If users wish to generate their own *.bam from other custom pipelines, make sure that:
- the UMI and cellbarcode information must be present in the *.bam file as custom tags CB:Z and UB:Z respectively, in this order.
- TS oligo sequence must keep intact on Read 1.
We recommend most users to run SCAFE using workflows with default options. There are 3 types of workflow: (1) "solo" for processing of a single library, (2) "aggregate" for aggregating of multiple libraries and (3) "subsample" for down-sampling a single library (for assessment of sequencing depth). "solo" accepts *.bam while "aggregate"/"subsample" accepts *.ctss.bed files (generated from tool.sc.bam_to_ctss). For multiple libraries, we recommend users to first run either workflow.sc.solo or tool.sc.bam_to_ctss on indiviudal libraries, and then take the *.ctss.bed file from all libraries to run workflow.cm.aggregate. Aggregating of libraries for defining tCRE is recommended because (1) it generally increases the sensitivity of tCRE detection and (2) it produces a common set of tCREs for all libraries so the IDs are portable between libraries. Please check the help messages for details of running the workflows:
#--- check out the help message of the three single cell workflows
scafe.workflow.sc.solo --help
scafe.workflow.cm.aggregate --help
scafe.workflow.sc.subsample --help
For the sake of flexibiity, SCAFE allows users to run individual tools with custom options for exploring the effect of cutoffs or supplying alternative intermediate inputs. See here for the full list of tools and their usage. Let's walk through a couple of individual tools with the demo data.
- tool.sc.bam_to_ctss: First, we convert a cellranger *.bam file to *.ctss.bed files. "ctss" refers as capped TSS, and *.ctss.bed file is a common format for storing TSS information. For procedural convenenice, tool.sc.bam_to_ctss generates multiple *.ctss.bed files at various levels of collapsing the signal (e.g. piling up UMI at TSS or not, summing up UMI of different cellbarcode or not). By default, the tool.sc.bam_to_ctss process ONLY primary alignments regardless of MAPQ. If users wants to, for example, include also secondary aligments with a minimum MAPQ (e.g. 10), the user could run as the followings:
#--- check out the help message of tool.sc.bam_to_ctss
scafe.tool.sc.bam_to_ctss --help
#--- run tool.sc.bam_to_ctss with custom options
scafe.tool.sc.bam_to_ctss \
--min_MAPQ 10 \
--exclude_flag '128,4' \
--overwrite=yes \
--bamPath=./demo/input/sc.solo/demo.cellranger.bam \
--genome=hg19.gencode_v32lift37 \
--outputPrefix=demo \
--outDir=./demo/output/sc.solo/bam_to_ctss/- tool.cm.remove_strand_invader: Then, we removes the strand invader artefacts from the *.ctss.bed file generated from tool.sc.bam_to_ctss. Please refer to here for the rationale of removing strand invader artefacts. If the users would like to use a more stringent cutoff to define strand invader artefacts, e.g. --min_edit_distance=3 and --min_end_non_G_num=1, so that less reads will be removed, and the user could run as the followings:
#--- check out the help message of tool.cm.remove_strand_invader
scafe.tool.cm.remove_strand_invader --help
#--- run tool.cm.remove_strand_invader with custom options
scafe.tool.cm.remove_strand_invader \
--min_edit_distance=3 \
--min_end_non_G_num=1 \
--overwrite=yes \
--ctss_bed_path=./demo/output/sc.solo/bam_to_ctss/demo/bed/demo.collapse.ctss.bed.gz \
--genome=hg19.gencode_v32lift37 \
--outputPrefix=demo \
--outDir=./demo/output/sc.solo/remove_strand_invader/- tool.cm.cluster: Then, we cluster the *.ctss.bed file into TSS clusters. Please refer to here for the rationale of clustering. By default, the clusters with <5 UMI within cluster, <3 UMI at summit or expressed in <3 cells were removed. If the users would like to use a more stringent cutoff to remove more lowly expressed TSS clusters, e.g. --min_cluster_count=10, --min_summit_count=5 and --min_num_sample_expr_cluster=5,the user could run as the followings:
#--- check out the help message of tool.cm.cluster
scafe.tool.cm.cluster --help
#--- run tool.cm.cluster with custom options
scafe.tool.cm.cluster \
--min_cluster_count=10 \
--min_summit_count=5 \
--min_num_sample_expr_cluster=5 \
--overwrite=yes \
--cluster_ctss_bed_path=./demo/output/sc.solo/bam_to_ctss/demo/bed/demo.collapse.ctss.bed.gz \
--outputPrefix=demo \
--outDir=./demo/output/sc.solo/cluster/- tool.cm.filter: Then, we filter the TSS cluster using logistic regression. By default, tool.cm.filter uses a pre-trained multiple logistic regression model from human iPSC cells using matched ATAC-Seq data. If the users would like to use their own matched ATAC-Seq data (–logP as *.bigwig file) for training of the regression model using --training_signal_path and --testing_signal_path options. Also, the user can set a permissive logistic probablity threshold (default=0.5) using --default_cutoff option (e.g. 0.3). The user could run as the followings:
#--- check out the help message of tool.cm.filter
scafe.tool.cm.filter --help
#--- run tool.cm.cluster with custom options
scafe.tool.cm.filter \
--default_cutoff=0.3 \
--overwrite=yes \
--ctss_bed_path=./demo/output/sc.solo/bam_to_ctss/demo/bed/demo.collapse.ctss.bed.gz \
--ung_ctss_bed_path=./demo/output/sc.solo/bam_to_ctss/demo/bed/demo.unencoded_G.collapse.ctss.bed.gz \
--tssCluster_bed_path=./demo/output/sc.solo/cluster/demo/bed/demo.tssCluster.bed.gz \
--training_signal_path=./demo/input/atac/demo.atac.bw \
--testing_signal_path=./demo/input/atac/demo.atac.bw \
--genome=hg19.gencode_v32lift37 \
--outputPrefix=demo \
--outDir=./demo/output/sc.solo/filter/- tool.cm.annotate: Finally, we will define and annotate tCREs based on the gene models in reference genome. By default, tool.cm.annotate merge TSS clusters located within +100nt and –400nt as a tCRE (defined by combination of option --CRE_extend_size and --CRE_extend_upstrm_ratio). Also, the tCREs with +/-500nt of annotated gene TSS will be assigned as proximal tCRE (defined by option --proximity_slop_rng). If the users would like to define tCRE by merging more distant TSS clusters (e.g. +200nt and –600nt) and assign tCRE further away (+/-1000nt) from gene TSS as proximal, the user could run as the followings:
#--- check out the help message of tool.cm.annotate
scafe.tool.cm.annotate --help
#--- run tool.cm.annotate with custom options
scafe.tool.cm.annotate \
--CRE_extend_size=800 \
--CRE_extend_upstrm_ratio=3 \
--proximity_slop_rng=1000 \
--overwrite=yes \
--tssCluster_bed_path=./demo/output/sc.solo/filter/demo/bed/demo.tssCluster.default.filtered.bed.gz \
--tssCluster_info_path=./demo/output/sc.solo/filter/demo/log/demo.tssCluster.log.tsv \
--genome=hg19.gencode_v32lift37 \
--outputPrefix=demo \
--outDir=./demo/output/sc.solo/annotate/Currently, four reference genomes ara available. See ./scripts/scafe.download.resources.genome for downloading. Alternatively, some users might work on genomes of other organisms, or prefer to use custom gene models for annotating tCREs. tool.cm.prep_genome converts user-supplied genome *.fasta and gene model *.gtf into necessary files for SCAFE. You can check out the help message for inputs of tool.cm.prep_genome and then test run a demo using TAIR10 genome with AtRTDv2 gene model.
#--- check out the help message of tool.cm.prep_genome
scafe.tool.cm.prep_genome --help
#--- run the tool on the TAIR10 assembly with gene model AtRTDv2
scafe.tool.cm.prep_genome \
--overwrite=yes \
--gtf_path=./demo/input/genome/TAIR10.AtRTDv2.gtf.gz \
--fasta_path=./demo/input/genome/TAIR10.genome.fa.gz \
--chrom_list_path=./demo/input/genome/TAIR10.chrom_list.txt \
--mask_bed_path=./demo/input/genome/TAIR10.ATAC.bed.gz \
--outputPrefix=TAIR10.AtRTDv2 \
--outDir=./demo/output/genome/SCAFE also accepts .*bam files from bulk CAGE. The major difference between singel cell and bulk workflow is cellbarcode is not considered. Otherwise, the options between single cell and bulk workflow are large the same. Please check the help messages for details of running the workflows:
#--- check out the help message of the three single cell workflows
scafe.workflow.bk.solo --help
scafe.workflow.cm.aggregate --help
scafe.workflow.bk.subsample --help- bam_to_ctss error check for various problems in bam file, e.g. bams with read 2 only
- enable the use of system-wide bins for common 3rd party software (e.g. bedtools samtools)
- update genome resources with updated genocode versions