ts-ipc provides a light-weight framework for end-to-end population genetic
simulations that utilize tree sequence recording to combine forward simulation
and coalescent simulation.
It facilitates communication between the forward simulator SLiM and python,
where much of the tree sequence ecosystem lives:
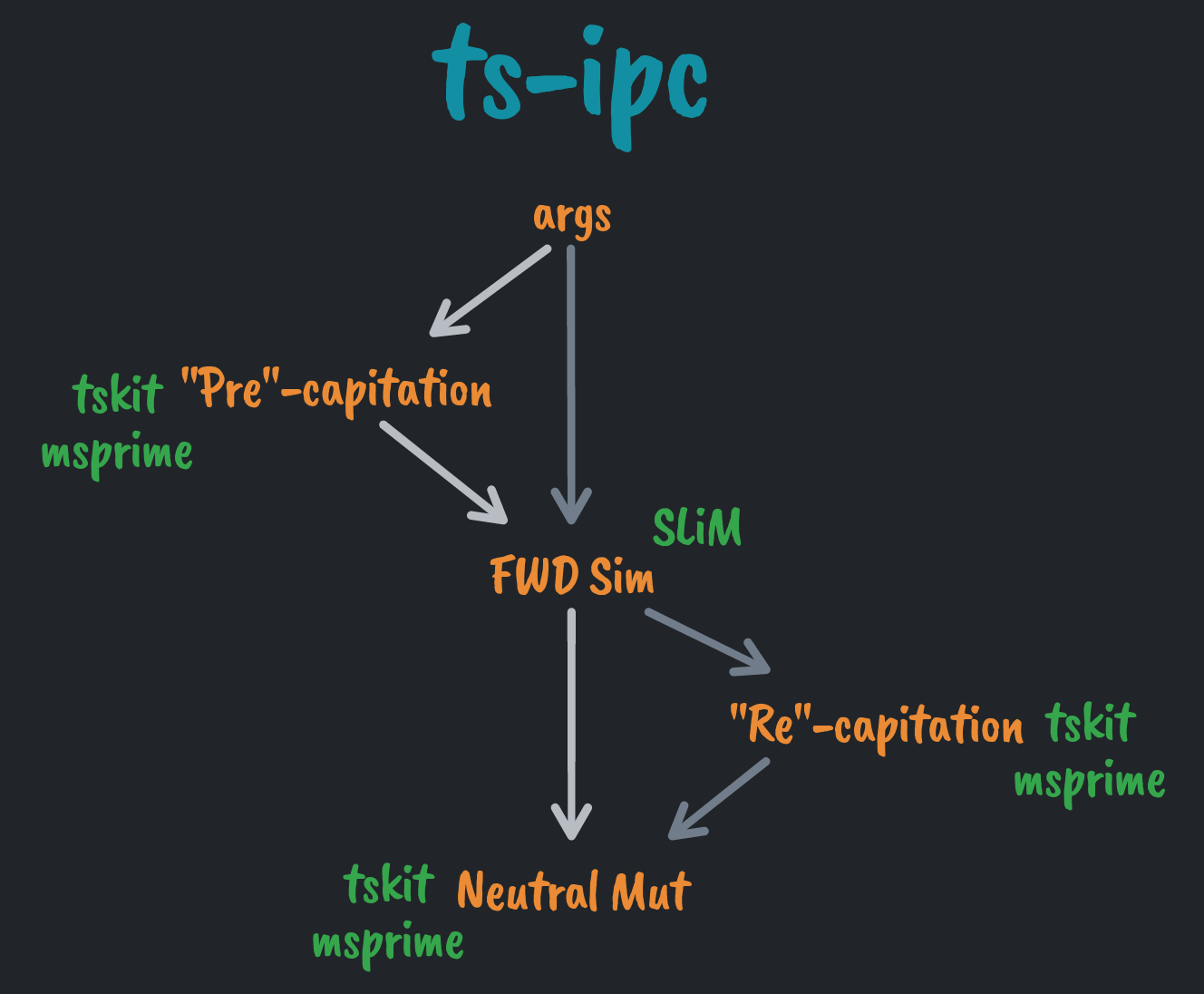
The major aim of this framework is to define one set of Arguments that will be
used by all simulation components. This is primarily for consistency: some
parameters may be shared between several components, so it is appealing to only
have to define them once.
Another aim is persistence: using the tree sequence data structure to save intermediate files, as well as to persist the entire simulation configuration into the tree sequence metadata allows for high reproducibility and easier debugging.
The framework provides various classes to either inherit from or use directly:
Arguments- based on python dataclasses, inherit from
Argumentsand define arguments for all simulation components in dataclass-style
- based on python dataclasses, inherit from
PreCapSim,Recapitation,AddNeutralMut- abstract base classes for python-based simulation components
- need to define the functions
_assert_requiredand_make_tsin custom child classes
FwdSim- run
SLiMsimulation, given.slimscript defined inArguments
- run
Simulation- define full simulation, given a set of components
For usage examples, see e.g.:
ArgumentsPreCapSimRecapitationAddNeutralMutFwdSimSimulation