🚧🚧 Please note that ConPLex is currently a pre-release and is actively being developed. For the code used to generate our PNAS results, see the manuscript code 🚧🚧
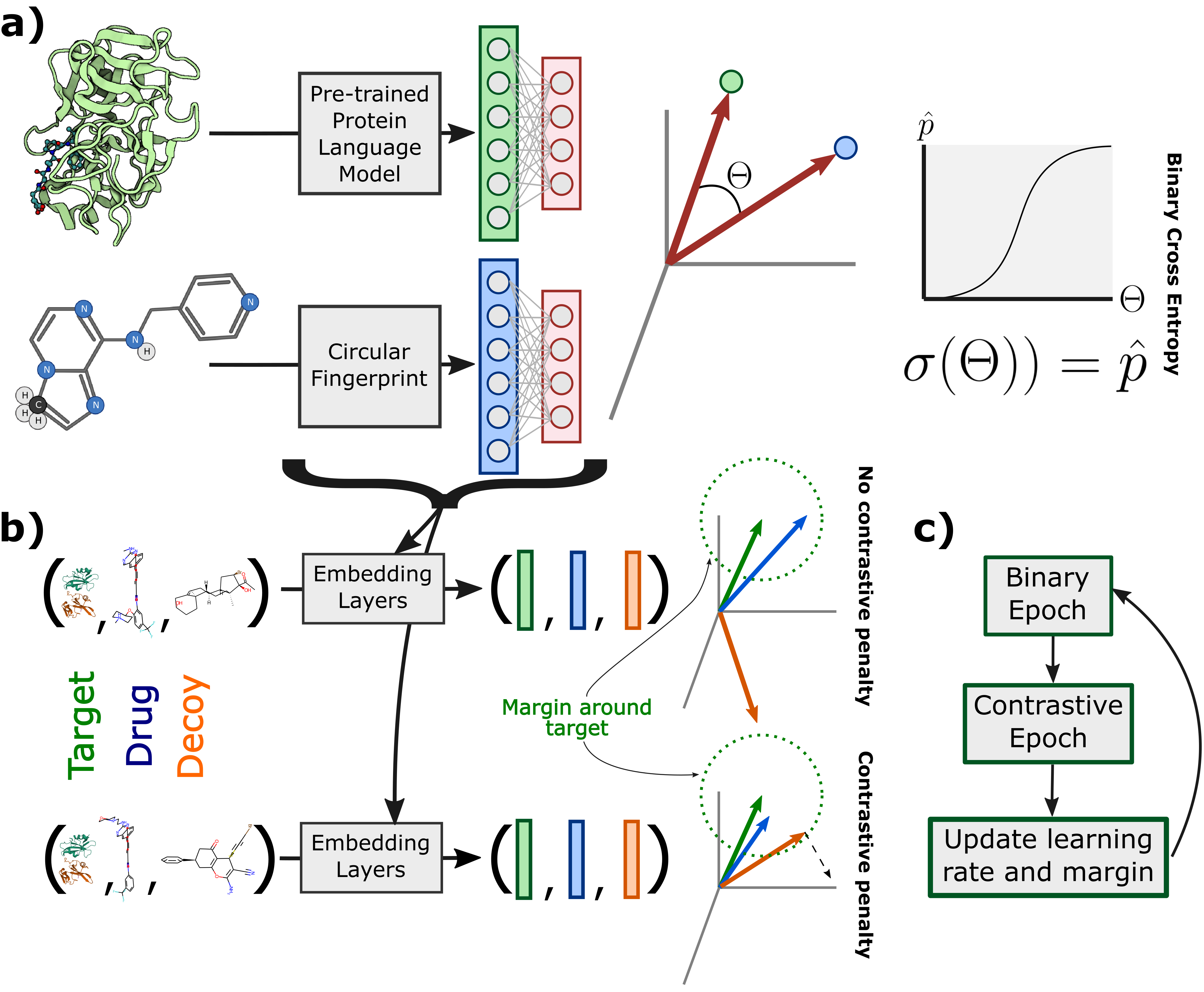
Sequence-based prediction of drug-target interactions has the potential to accelerate drug discovery by complementing experimental screens. Such computational prediction needs to be generalizable and scalable while remaining sensitive to subtle variations in the inputs. However, current computational techniques fail to simultaneously meet these goals, often sacrificing performance on one to achieve the others. We develop a deep learning model, ConPLex, successfully leveraging the advances in pre-trained protein language models ("PLex") and employing a novel protein-anchored contrastive co-embedding ("Con") to outperform state-of-the-art approaches. ConPLex achieves high accuracy, broad adaptivity to unseen data, and specificity against decoy compounds. It makes predictions of binding based on the distance between learned representations, enabling predictions at the scale of massive compound libraries and the human proteome. Experimental testing of 19 kinase-drug interaction predictions validated 12 interactions, including four with sub-nanomolar affinity, plus a novel strongly-binding EPHB1 inhibitor (
You should first have a version of cudatoolkit compatible with your system installed. Then run
pip install conplex-dti
conplex-dti --helpgit clone https://github.com/samsledje/ConPLex.git
cd ConPLex
conda create -n conplex-dti python=3.9
conda activate conplex-dti
make poetry-download
# If this fails at rdkit, just pip install rdkit==2022.9.5
export PATH="[poetry install location]:$PATH"
export PYTHON_KEYRING_BACKEND=keyring.backends.null.Keyring
make install
conplex-dti --help
# Extra dependencies
pip install pyfaidx==0.8.1.1conplex-dti download --to datasets --benchmarks davis bindingdb biosnap biosnap_prot biosnap_mol dudeconplex-dti download --to . --models ConPLex_v1_BindingDBconplex-dti train --run-id TestRun --config config/default_config.yamlconda install -c conda-forge ncbi-datasets-cli
mkdir -p datasets/genomes
# Download
datasets download genome taxon human --reference --assembly-level chromosome --include protein --dehydrated --filename datasets/genomes/human.zip
datasets download genome taxon fungi --reference --assembly-level chromosome --include protein --dehydrated --filename datasets/genomes/fungi.zip
datasets download genome taxon bacteria --reference --assembly-level complete --include protein --dehydrated --filename datasets/genomes/bacteria.zip
# Unzip
unzip datasets/genomes/human.zip -d datasets/genomes/human
unzip datasets/genomes/fungi.zip -d datasets/genomes/fungi
unzip datasets/genomes/bacteria.zip -d datasets/genomes/bacteria
# Rehydrate
datasets rehydrate --directory datasets/genomes/human
datasets rehydrate --directory datasets/genomes/fungi
datasets rehydrate --directory datasets/genomes/bacteria
# Clean
mkdir -p datasets/proteomes
python process_faa.py --files datasets/genomes/human/ncbi_dataset/data/*/protein.faa --output datasets/proteomes/human.tsv
python process_faa.py --files datasets/genomes/fungi/ncbi_dataset/data/*/protein.faa --output datasets/proteomes/fungi.tsv
python process_faa.py --files datasets/genomes/bacteria/ncbi_dataset/data/*/protein.faa --output datasets/proteomes/bacteria.tsvconplex-dti embed --moltype [protein or molecule] --data-file [protein seqs or molecule SMILES].tsv --model-path ./models/ConPLex_v1_BindingDB.pt --outfile ./results.npzFormat of [pair predict file].tsv should be [protein ID]\t[molecule ID]\t[protein Sequence]\t[molecule SMILES]
conplex-dti predict --data-file [pair predict file].tsv --model-path ./models/ConPLex_v1_BindingDB.pt --outfile ./results.tsv# Install
pip install chromadb
# Download into ./dbs folder from box# Use chroma to query protein coembeddings with molecule coembeddings
import numpy as np
import chromadb
molecule_embeddings = np.load('natural_products_embeddings.npz', allow_pickle=True)
mols = molecule_embeddings['embedding'].tolist()
client = chromadb.PersistentClient(path="./dbs")
collection = client.get_or_create_collection(name="conplex_v0", metadata={"hnsw:space": "cosine"})
results = collection.query(
query_embeddings=mols,
n_results=50,
)results is a dictionary of lists containing n_results return values for each query with the structure
{
'ids': [[str]],
'documents': [[protein seqs: str]]
'metadatas': [[
{
'name': str,
'kingdom: str
},
]],
'distances': [[cosine similarity: float]]
}If you use ConPLex, please cite Contrastive learning in protein language space predicts interactions between drugs and protein targets by Rohit Singh*, Samuel Sledzieski*, Bryan Bryson, Lenore Cowen and Bonnie Berger.
@article{singh2023contrastive,
title={Contrastive learning in protein language space predicts interactions between drugs and protein targets},
author={Singh, Rohit and Sledzieski, Samuel and Bryson, Bryan and Cowen, Lenore and Berger, Bonnie},
journal={Proceedings of the National Academy of Sciences},
volume={120},
number={24},
pages={e2220778120},
year={2023},
publisher={National Acad Sciences}
}Thanks to Ava Amini, Kevin Yang, and Sevahn Vorperian from MSR New England for suggesting the use of the triplet distance contrastive loss function without the sigmoid activation. The default has now been changed. For the original formulation with the sigmoid activation, you can set the --use-sigmoid-cosine flag during training.
Code used to generate results in the manuscript can be found in the development repository