The Extracellular Recording Site Simulator (ERSS) is a tool for creating biophysically realistic recording sites given a probe geometry and brain region. EERSS currently supports the Neuropixels probe geometry and two detailed Somatosensory neocortical volumes from rat studies.
To get started with ERSS, clone the repo into your code base.
git clone https://github.com/colehurwitz31/ERSS.gitTo create a new cortex object, choose a brain region (currently, only the cortex region is supported) and import the class from that region's folder.
Once you have imported the class from region folder, you can decide which dataset and animal data to use to reconstruct the region. Currently, I have implemented a rat barrel cortex reconstruction using data from the Helmstaedter series of papers from 2010-2011.
from regions.cortex.CortexColumn import Column
from regions.cortex.rat_data.CortexDataDicts import createHelmstaedterRatSomatosensoryCortexDataDict
rat_somatosensory_cortex_data_dict = createHelmstaedterRatSomatosensoryCortexDataDict()
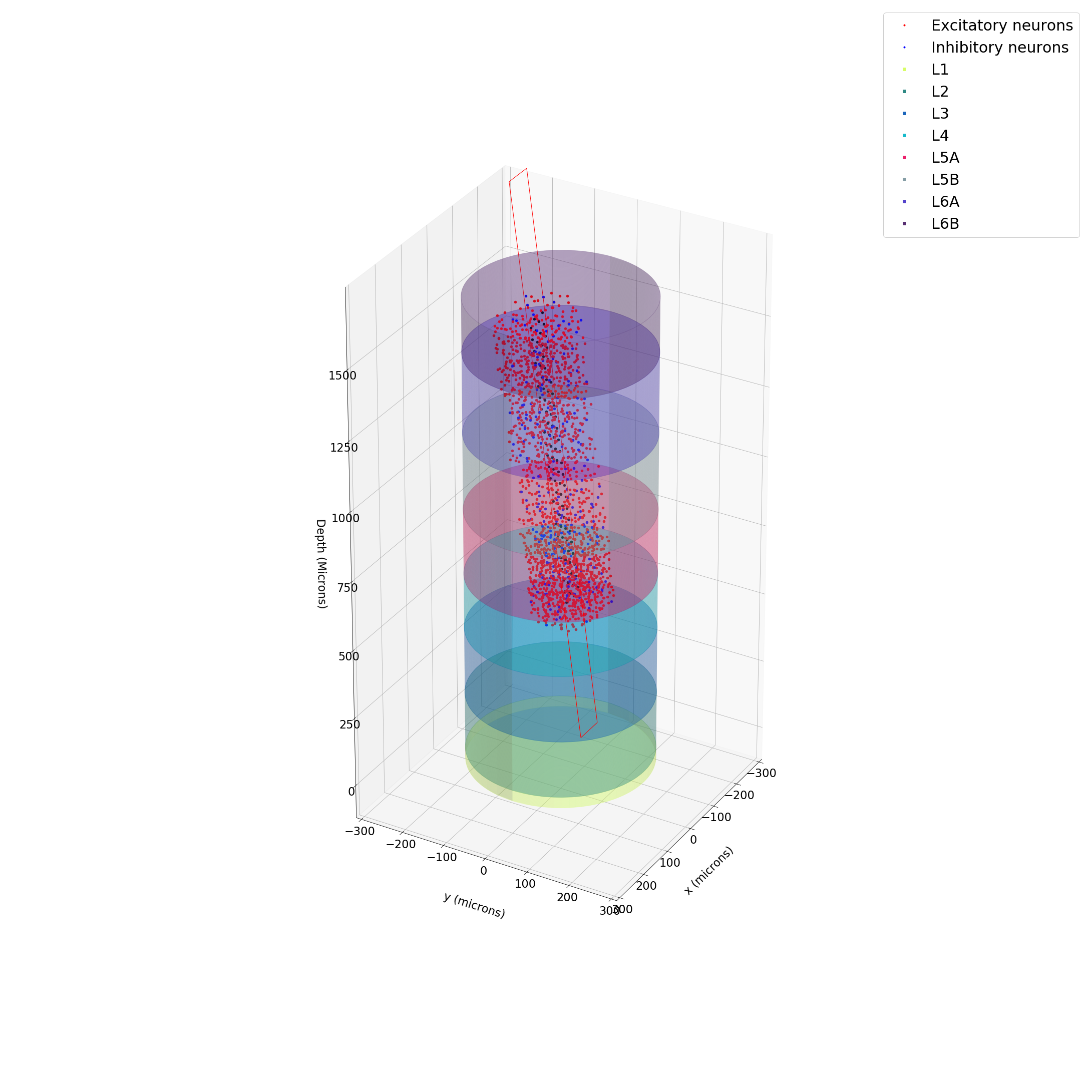
column = Column(rat_somatosensory_cortex_data_dict, radius=200.0, min_neuron_dist=15)Each column class is constructed by inputting a radius (microns) and minimum distance (microns) between neurons and then randomly sampling from a mean and std devation for each attribute required for the reconstruction. After construction, the neuron and layer information is stored in pandas DataFrames. You can easily plot the whole column or specific layers to see the neuronal layout.
layer_id = 1
fig= plt.figure(figsize=(10,10))
ax = Axes3D(fig, azim=30, elev=30)
column.plotLayer(layer_id, ax=ax)Finally, you can construct a probe object by importing a premade probe from the PremadeProbes class in the probes folder. Currently, only Neuropixels is supported, but other probes will be added in future updates. You can specify the number of channels on the probe in the construction (the default insertion point of the probe is at the origin of the plot which is the central surface of the barrel cortex.
from ERSS.probes.PremadeProbes import NeuropixelsProbe
probe = NeuropixelsProbe(num_channels = 192)Probes store the channel information of the probe in a pandas DataFrame and can be rotated and shifted to make for more realistic insertions into the brain region. Once you are satisfied with the probe position (you can draw the probe and the column to see their relative positions), you can get the neurons within a radius (microns) around the channels of your choosing.
You can then plot the neurons within that radius to see how the recording site would look for a probe.
close_neuron_df = probe.getNeuronsRadius(column.getNeurons(), radius=80.0, excluded_channel_ids=excluded_channel_ids)
fig=plt.figure(figsize=(30,30))
ax = Axes3D(fig, azim=30, elev=30)
probe.draw(ax, excluded_channel_ids=excluded_channel_ids)
column.plotColumn(close_neuron_df, plot_neurons=True, ax=ax)- Neuropixels
-
Somatosensory neocortical volume from HelmStaedter papers
-
Somatosensory neocortical volume from BlueBrain portal
This software was designed as an initial step in creating brain/probe specific evaluation datasets for spike sorting.
It can be used to generate realistic extracellular recording sites for different probe geometries within different brain regions that can be simulated using various extracellular simulators such as ViSAPy and LFPy.
I will implement more probe types (Neuroseeker, H-Series, tetrodes, etc.) along with more brain regions (hippocampus, thalamus, etc.) and animals (mouse).
Meyer, Hanno S., et al. "Inhibitory interneurons in a cortical column form hot zones of inhibition in layers 2 and 5A." Proceedings of the National Academy of Sciences 108.40 (2011): 16807-16812. APA
Meyer, Hanno S., et al. "Number and laminar distribution of neurons in a thalamocortical projection column of rat vibrissal cortex." Cerebral cortex 20.10 (2010): 2277-2286.
Meyer, Hanno S., et al. "Cell type–specific thalamic innervation in a column of rat vibrissal cortex." Cerebral cortex 20.10 (2010): 2287-2303.
Cole Hurwitz - The Institute for Adaptive and Neural Computation (ANC), University of Edinburgh, Edinburgh, Scotland
For any correspondence, contact Cole Hurwitz at colehurwitz@gmail.com