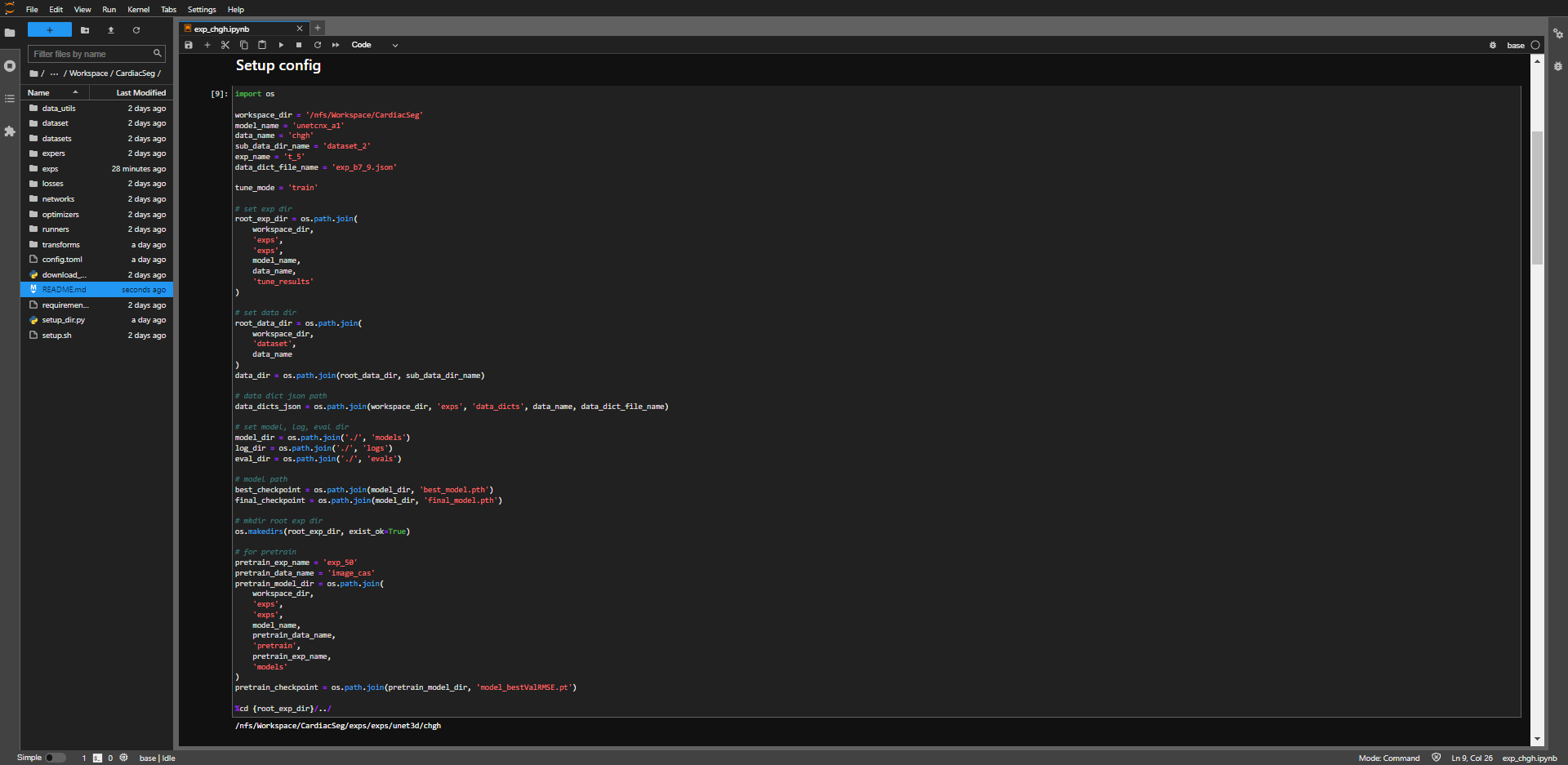
- setup data and model dir.
- set config to
config.toml, for download data and model.
# dataset google dirve file id
[dataset]
chgh='<google_drive_id>'
mmwhs='<google_drive_id>'
# model google dirve file id
[model.chgh]
unet3d='<google_drive_id>'
attention_unet='<google_drive_id>'
cotr='<google_drive_id>'
unetr='<google_drive_id>'
swinunetr='<google_drive_id>'
unetcnx_a1='<google_drive_id>'
[model.mmwhs]
unet3d='<google_drive_id>'
attention_unet='<google_drive_id>'
cotr='<google_drive_id>'
unetr='<google_drive_id>'
swinunetr='<google_drive_id>'
unetcnx_a1='<google_drive_id>'
- open training notebook from
CardiacSeg/exps/exp_chgh.ipynb.
- setup absolute path of workspace.
workspace = '<workspace>/CardiacSeg'
- setup model name.
- The model name used in this study is
unetcnx_a1.
- If you want to replace it with other research methods, you can change it to a different model name, such as
swinunetr, unetr, cotr, attention_unet and unet.
model_name = 'unetcnx_a1'
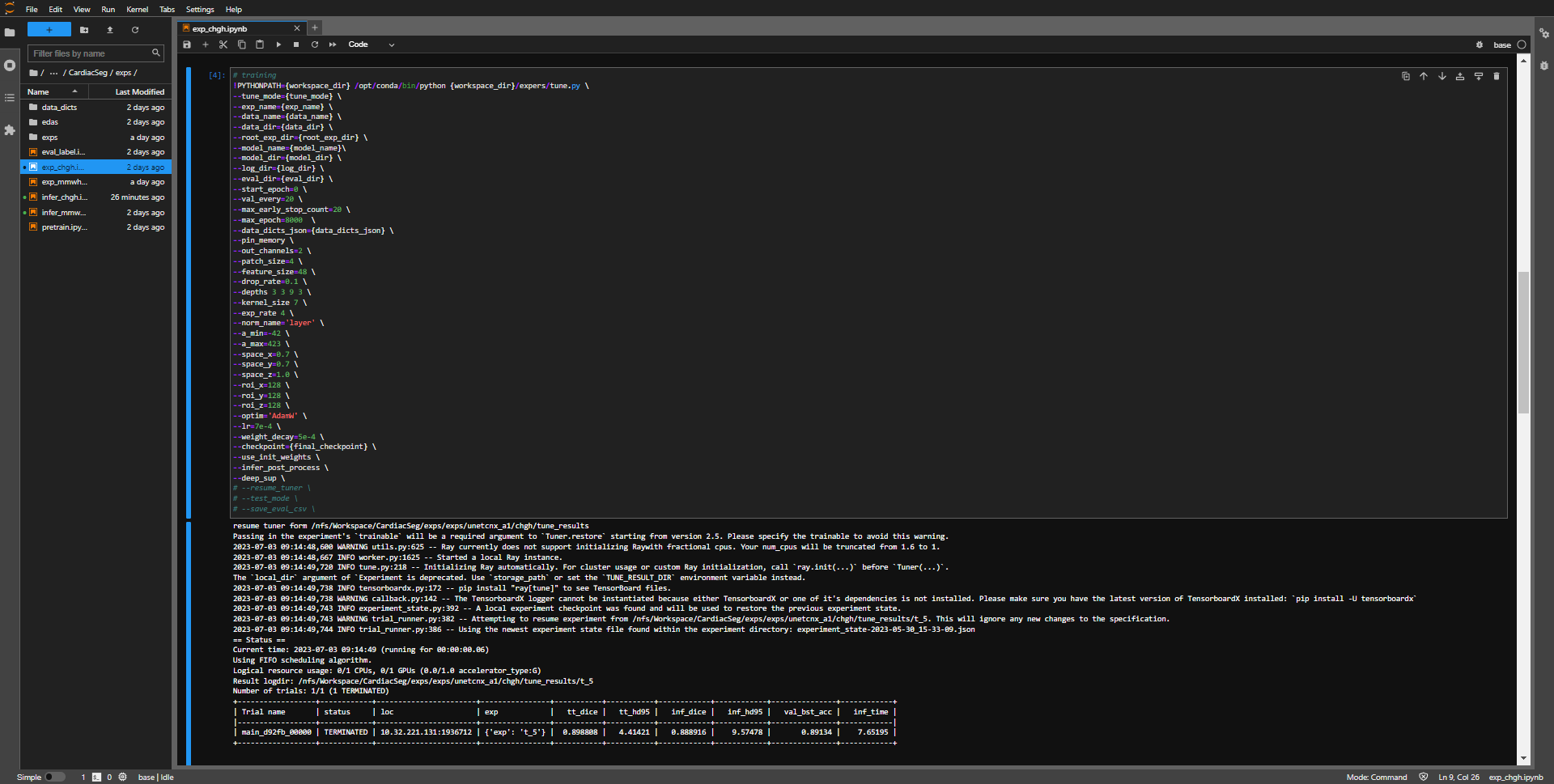
- run all cells, and the final results of the program will display validation scores and inference scores.
- open training notebook from
CardiacSeg/exps/infer_chgh.ipynb.
- setup absolute path of workspace.
workspace = '<workspace>/CardiacSeg'
- setup model name.
- The model name used in this study is
unetcnx_a1.
- If you want to replace it with other research methods, you can change it to a different model name, such as
swinunetr, unetr, cotr, attention_unet and unet.
model_name = 'unetcnx_a1'
- setup model name.
- If you want to replace it with other research methods, such as
swinunetr, unetr, cotr, attention_unet and unet. you can change it to a different exp name 't_4'.
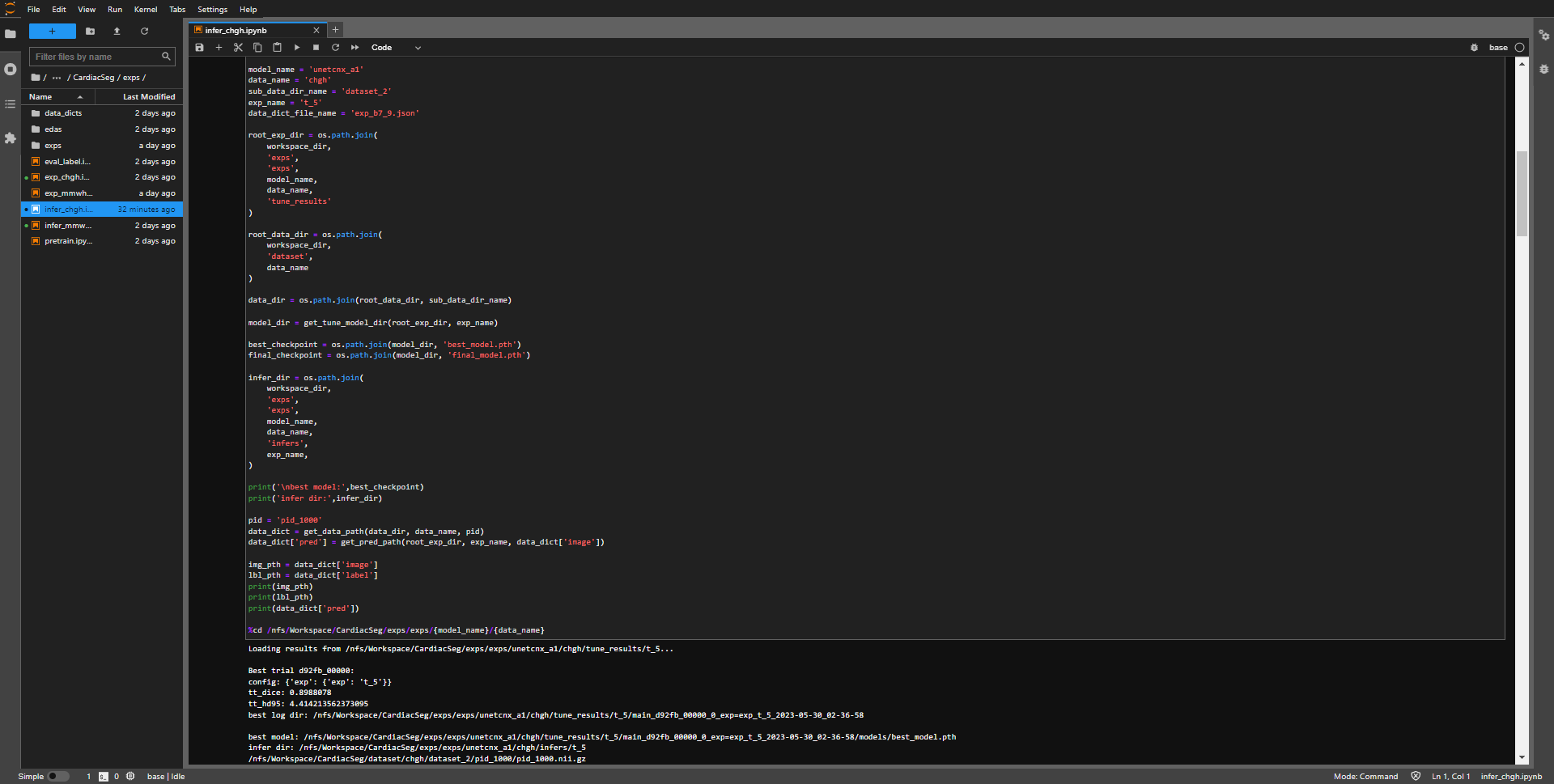
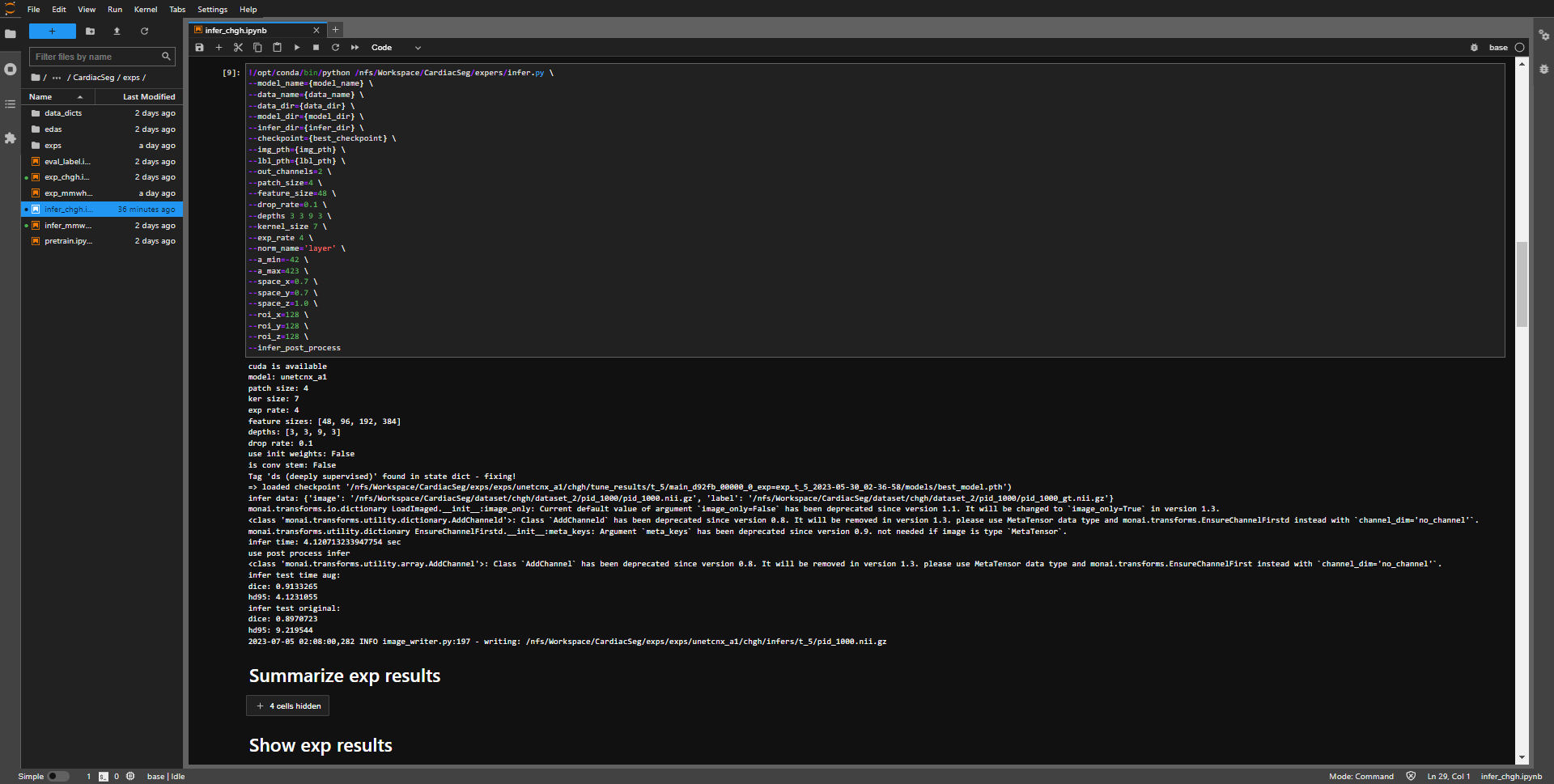
- after the inference is completed, the program will output the inference result and display the path of the output result (last line).
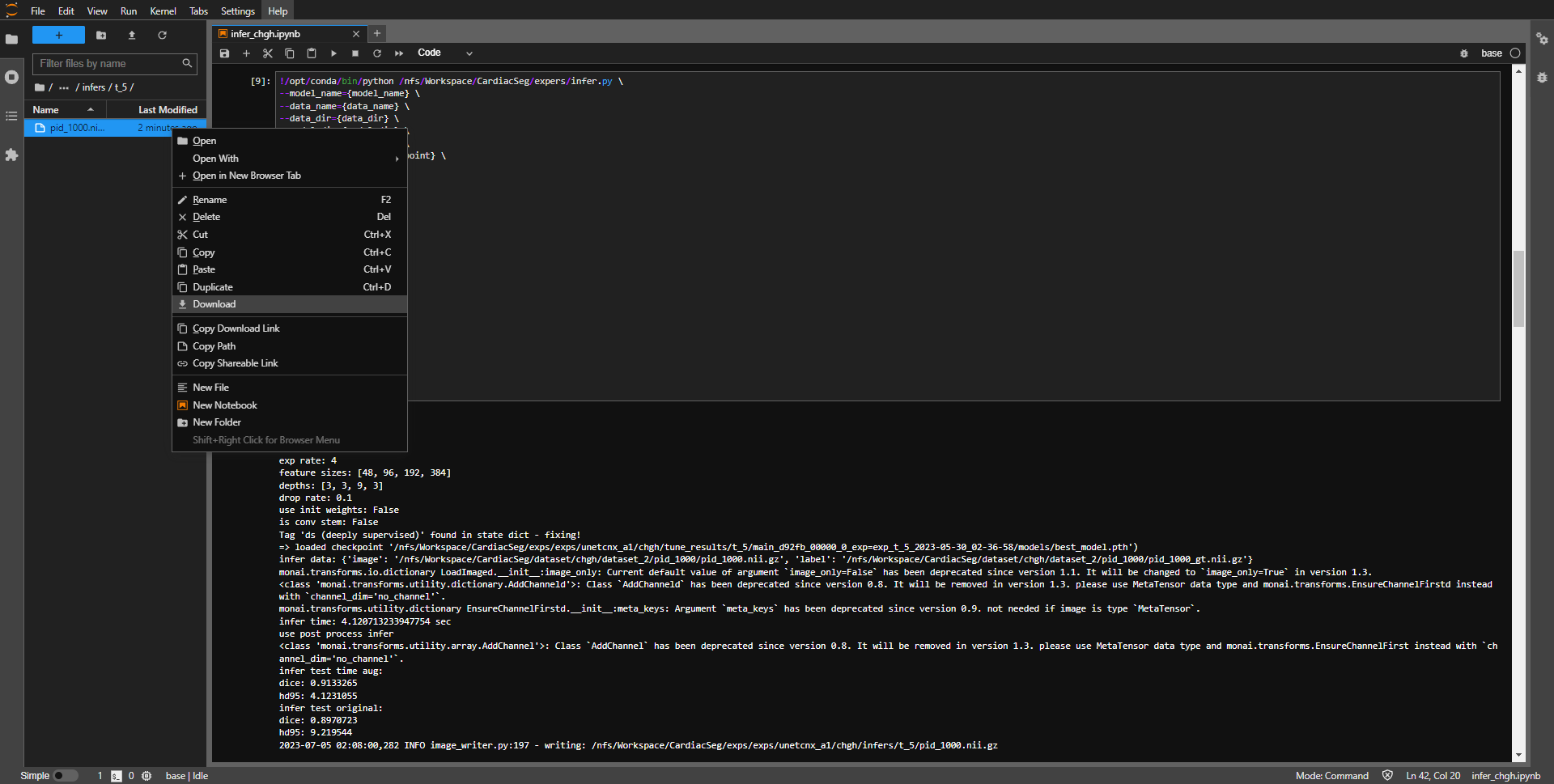
- download inference result
- display the inference results using 3D Slicer.