Quantifying the Effects of Contact Tracing, Testing, and Containment Measures in the Presence of Infection Hotspots
This repository contains the code base to run the sampling algorithm of a high-resolution spatiotemporal epidemic model at large scale, which can be used to predict and analyze the spread of epidemics such as COVID-19 in any real-world city and region. Different testing & tracing strategies, social distancing measures and business restrictions can be employed, amended, and extended arbitrarily in a modular fashion. Details about the relevant theory and methods can be found in our paper.
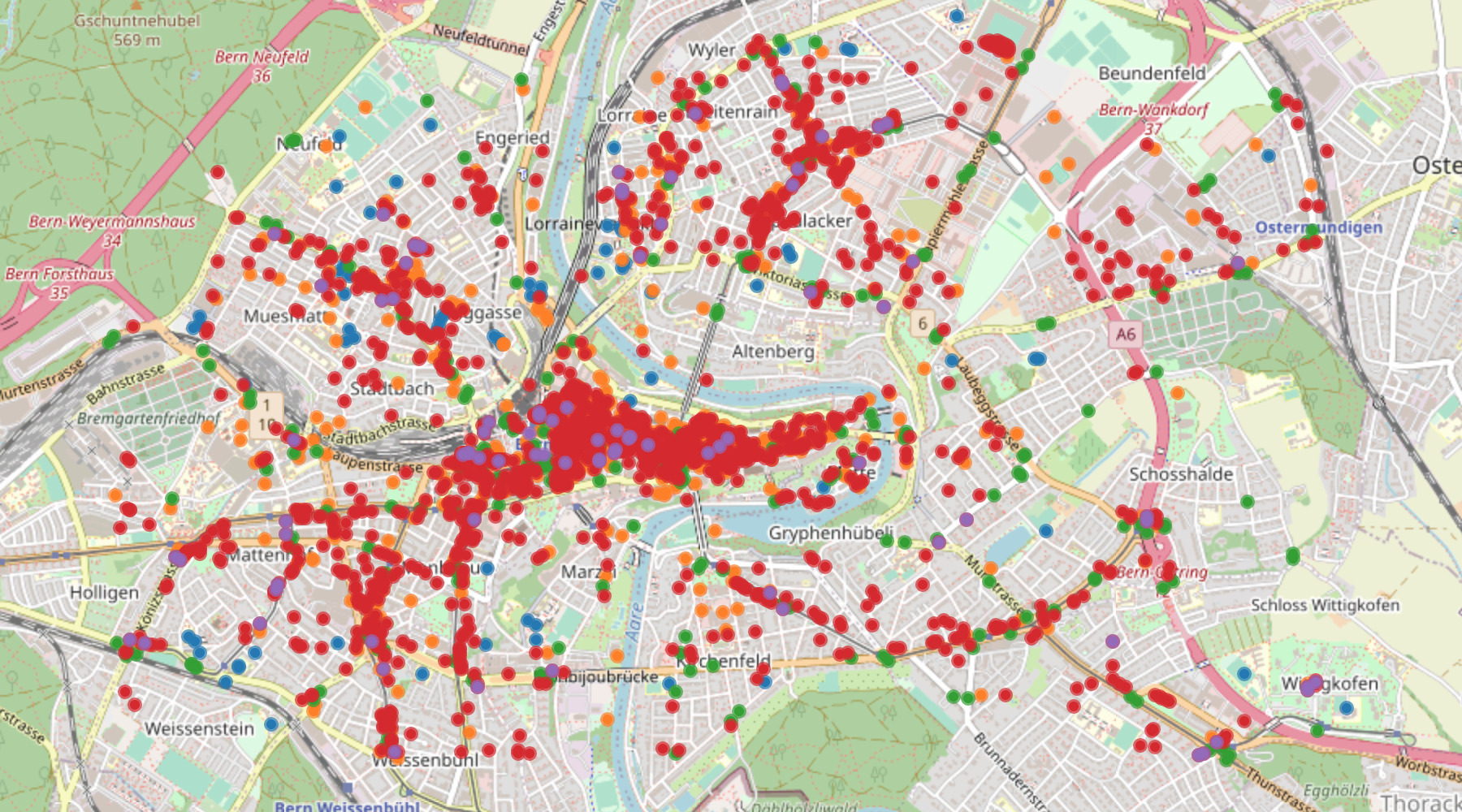
Motivated by several lines of evidence suggesting for superspreading events or infection hotspots to play a key role in the transmission dynamics of COVID-19, we introduce an epidemiological modeling framework that explicitly represents visits to sites where infections occur or hotspots may emerge. We use temporal point processes to represent the state of each individual over time with respect to their mobility patterns, health, and testing status. The model leverages the locations of publicly available data on real-world sites and population density in a heuristic mobility model of a given region (below: Bern, Switzerland), whose assumptions can be arbitrarily generalized.
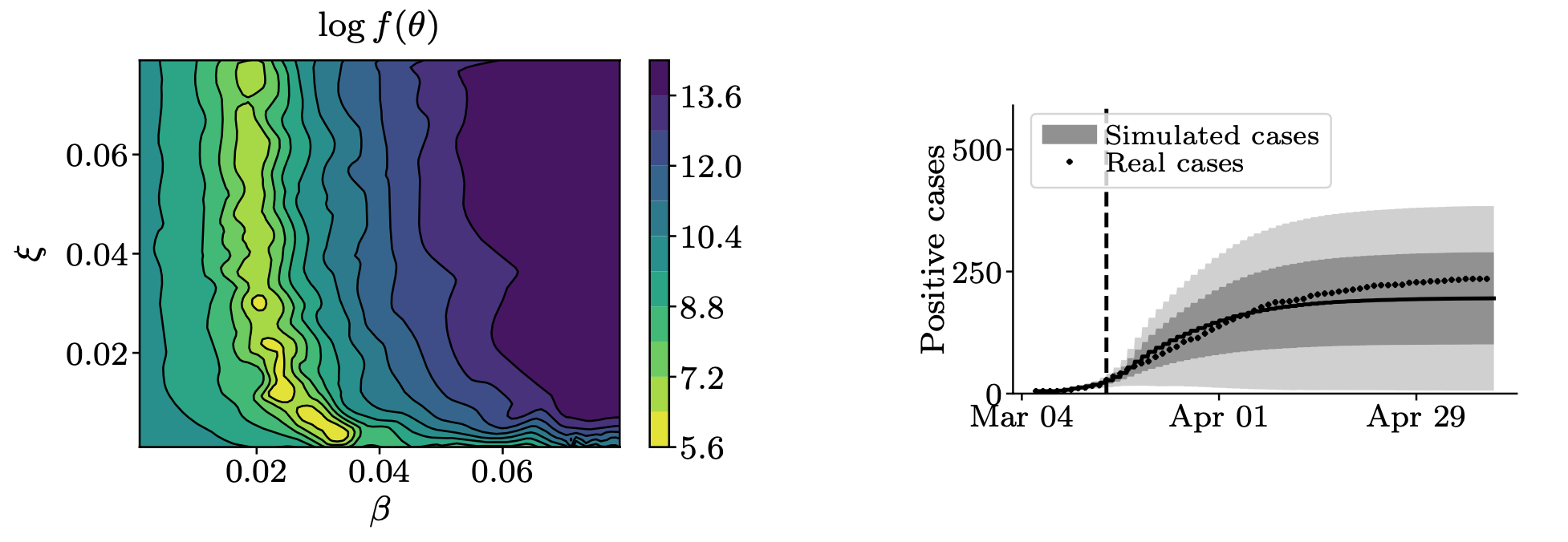
Bayesian optimization is used to estimate mobility-related exposure parameters by fitting the model to true observed case counts in a considered area. In our work on COVID-19, we study several regions in Germany and Switzerland, whose models were fit to real case counts over two-month windows before and during the "lockdown" in spring 2020:
Using the estimated parameters of the region-specific models, the sampling algorithm allows for analyses of counterfactual scenarios under various local circumstances.
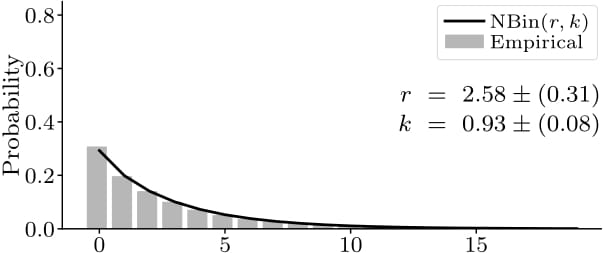
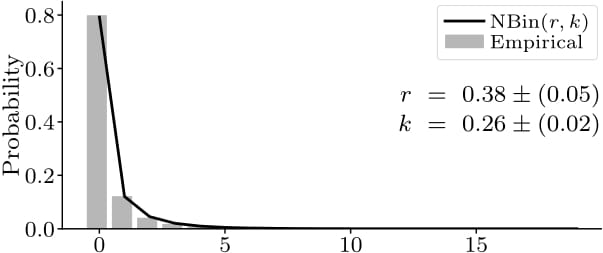
Contrary to existing agent-based epidemiological models, under our framework, the number of infections caused by infected individuals naturally emerges to be overdispersed, i.e., exhibiting greater variance than expected under the common Poisson assumption (here: fitted negative binomial distribution of the secondary infections overall and during a single site visit, respectively).
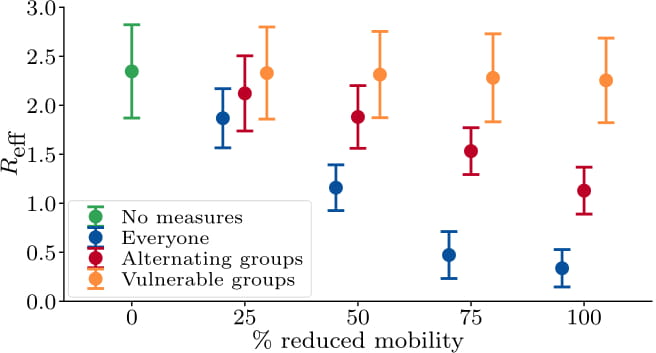
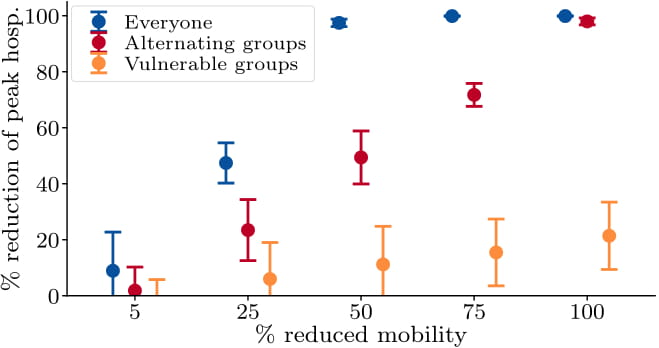
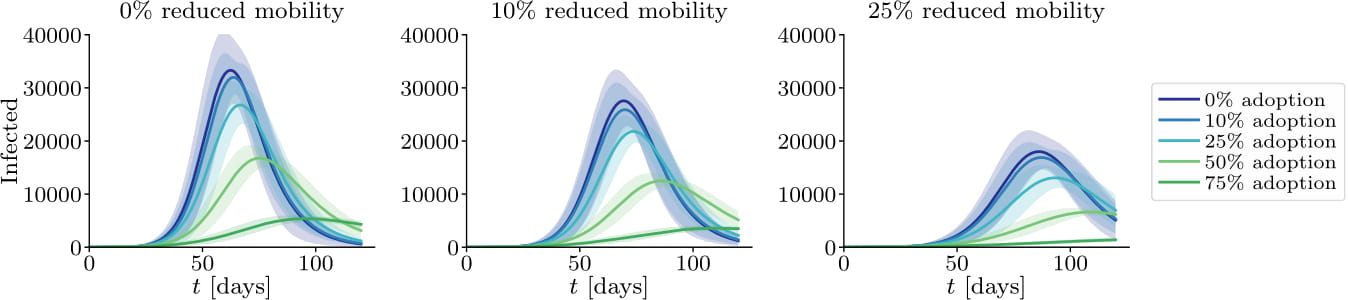
Amongst several other things, this framework allows for studying the effects of: the effective reproduction number, infections, hospitalizations, and fatalities over time during, e.g., mobility reduction, contact tracing, and testing measures, targeting not only the broader population, but also specific sites or individuals.
The sim/ directory contains the entire project code, where all simulator-specific code is situated inside sim/lib/. The simulator operates using the following main modules and rough purpose descriptions:
sim/lib/ |
Description |
|---|---|
| dynamics.py | Simulator core; defines a DiseaseModel object for simulating the spread of the epidemic. |
| measures.py | Containement measures; defines a Measure object for implementing intervention policies. |
| mobilitysim.py | Mobility patterns; defines a MobilitySimulator object for mobility traces of individuals. |
| distributions.py | Epidemiology; contains COVID-19 constants and distribution sampling functions. |
| calibrationFunctions.py; calibrationParser.py; calibrationSettings.py | Parameter Estimation; defines Bayesian Optimization pipeline for estimating region-specific exposure parameters |
| experiment.py | Analysis; defines an Experiment object for structured analysis and plotting of scenarios |
| plot.py | Plotting; defines a Plotter object for generating plots. |
The sim/ directory itself containts several scripts of the form sim-*.py, which run experiments and simulations as reported in our paper parallelized across CPUs. To execute a specific experiment script for an already fitted region, simply execute e.g. sim-*.py --country GER --area TU (here: Tübingen, Germany).
To apply the entire framework for a new region in experiments as defined in sim-*.py, the following two major steps need to be performed in order beforehand and only once:
- Create a new mobility file using the
sim/town-generator.ipynbnotebook, which fixes the region-specific metadata for simulation and stores it insidesim/lib/mobility. A downsampling factor for population and sites can be used to speed-up the simulation initially or for parameter estimation. The directory already contains all mobility metadata files we used in our simulations. - Estimate the region-specific exposure parameters by executing
calibrate.py. Before doing so, add the mobility file path, a region-specific code (e.g.GERandTU), and other details from above tosim/lib/calibrationSettings.py, following the structure for the existing regions. Hyperparameters for parameter estimation can be set using the command line arguments specified insim/lib/calibrationParser.py, for example as listed insim/lib/settings/estimation.md. The estimated parameters are saved and logged insidesim/logs. Depending on the size of the model, this step represents the major computational (yet one-time) cost.
Thus, the region metadata file in sim/lib/mobility and the parameter estimation log in sim/logs represents the fixed state of a corresponding region, starting point for simulating scenarios and running counterfactual analyses.
The results of a set of simulations such as sim-*.py are stored inside sim/summaries/ and can be visualized using sim/sim-plot.ipynb.
If you use parts of the code in this repository for your own research purposes, please consider citing:
@article{lorch2022quantifying,
title={Quantifying the Effects of Contact Tracing, Testing, and Containment Measures in the Presence of Infection Hotspots},
author={Lars Lorch and Heiner Kremer and William Trouleau and Stratis Tsirtsis and Aron Szanto and Bernhard Sch{\"o}lkopf and Manuel Gomez-Rodriguez},
journal={ACM Transactions on Spatial Algorithms and Systems},
year={2022}
}
This project uses Python 3. To create a virtual environment and install the project dependencies, you can run the following commands:
python3 -m venv env
source env/bin/activate
pip install -r requirements.txt