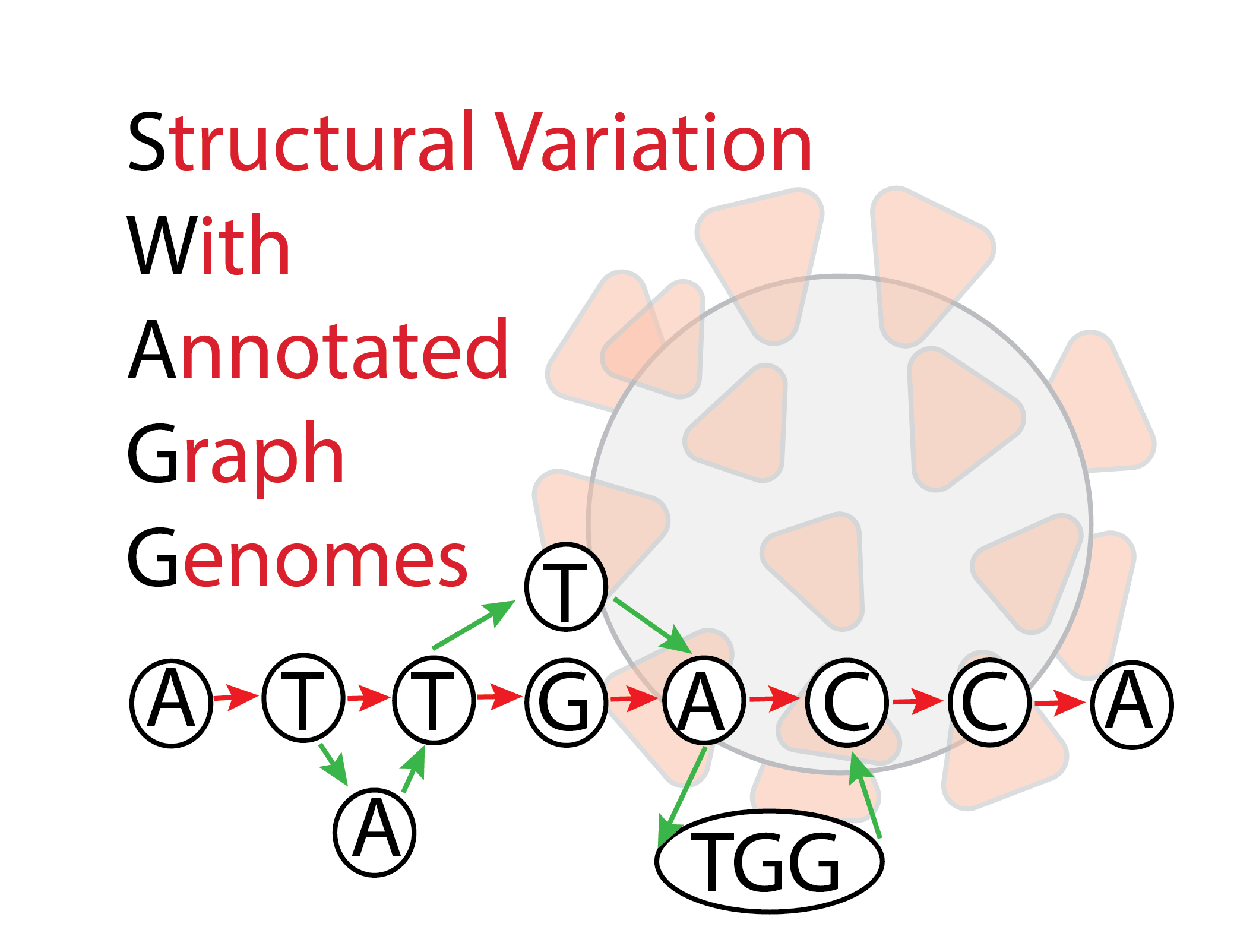
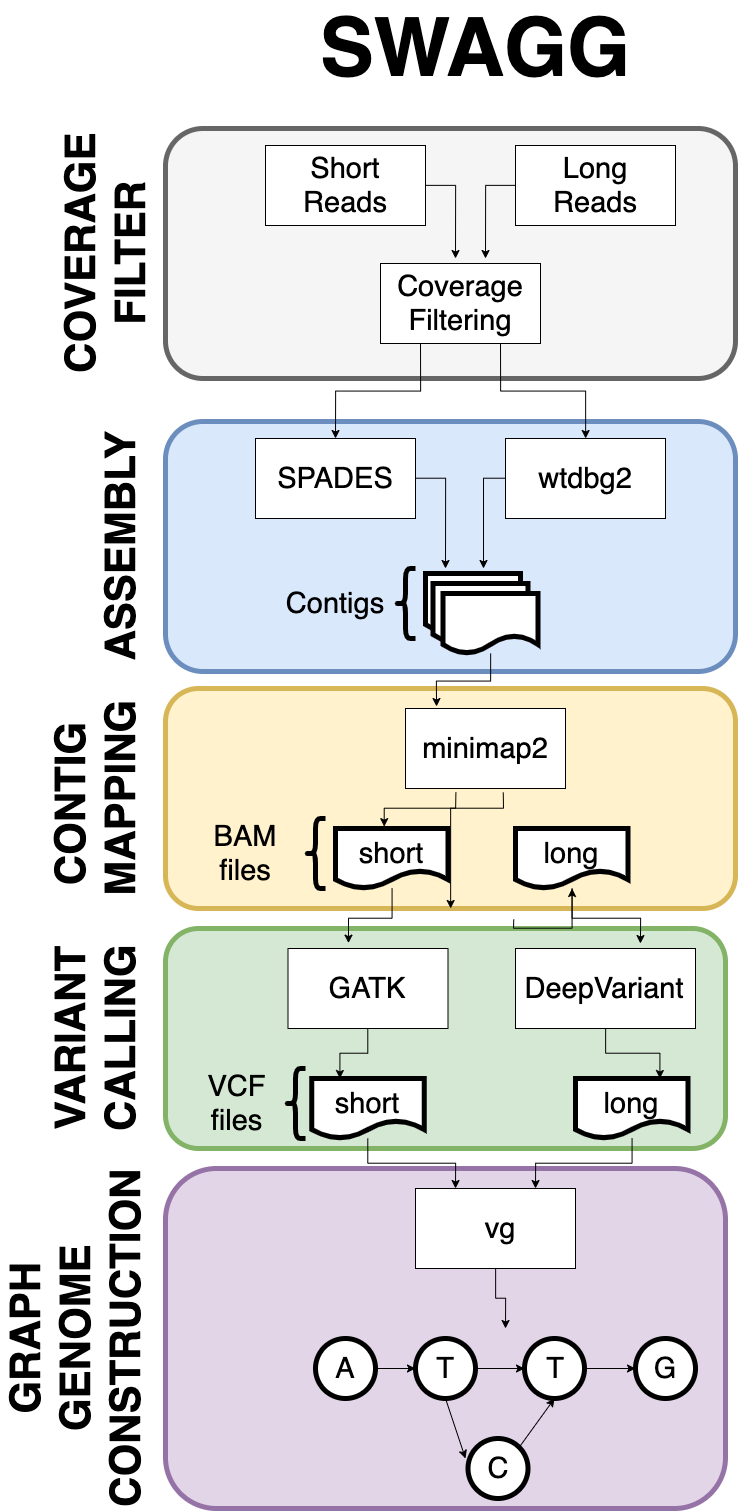
The SWAGG Pipeline is is designed to create graph genomes from read data. The input into the pipeline is a set of reads with a reference genomes, and the reads can be short reads or preprocessed long reads. Thereafter the reads are sent through a stage where they are assembled into longer contigs, and the resultant contigs are mapped back to the reference genome to look for discrepancies with the reference genome. These "discrepancies" or mutations are found using structural variant tools that output VCF files per read set. These VCF files are taken together to contrstruct the genome graph at the end of the pipeline.
Overview Diagram
We provide two options for installing : Docker or directly from Github.
The Docker image contains as well as a webserver and FTP server in case you want to deploy the FTP server. It does also contain a web server for testing the main website (but should only be used for debug purposes).
docker pull ncbihackathons/<this software>command to pull the image from the DockerHubdocker run ncbihackathons/<this software>Run the docker image from the master shell script- Edit the configuration files as below
git clone https://github.com/NCBI-Hackathons/<this software>.git- Edit the configuration files as below
sh server/<this software>.shto test- Add cron job as required (to execute .sh script)
Examples here
We tested four different tools with . They can be found in server/tools/ .
comes with a Dockerfile which can be used to build the Docker image.
git clone https://github.com/NCBI-Hackathons/<this software>.gitcd serverdocker build --rm -t <this software>/<this software> .docker run -t -i <this software>/<this software>
There is also a Docker image for hosting the main website. This should only be used for debug purposes.
git clone https://github.com/NCBI-Hackathons/<this software>.gitcd Websitedocker build --rm -t <this software>/website .docker run -t -i <this software>/website