The goal of this project is to provide a simple, hackable implementation of protein transformer models for education and research. Inspired by Karpathy's simple, hackable approach in the makemore series.
In this series of video lectures, we build a protein transformer from scratch and walk through the training and evaluation described below
- Protein transformers from scratch (5 lectures)
There are three main sections, all implemented in PyTorch:
- protein data loaders and tokenizers
- models ("decoder only" transformer)
- training loop
The provided ProteinDataset uses a character-level tokenizer (as used by ESM and all other protein language models I know about) and is defined in data.py. An example of using the ProteinDataset class:
proteins = [
"MCLLSLAAATVAARRTPLRLLGRGLAAAMSTAGPLKSV",
"MSSQIKKSKTTTKKLVKSAPKSVPNAAADDQIFCCQFE",
"MCLLSLAAATVAARRTPLRLLGRGLAAAMSTAGPLKSV",
]
chars = "ACDEFGHIKLMNPQRSTVWY"
max_length = 38
dataset = ProteinDataset(proteins, chars, max_length)The model implemented here is a standard "decoder-only" transformer architecture ("decoder-only" meaning that we use a triangular mask and ask the model to predict the next token). Following makemore, the implementation is totally spelled out in Python code so we can see all the details.
config = ModelConfig(vocab_size=20, block_size=128,
n_layer=4, n_head=4, embed_dim=64)
model = Transformer(config)In the training loop portion of the code, we parse the arguments from the command line, and then create the Adam optimizer, model instantiation, and proceed to train the model via backprop.
Every so often, we evaluate the loss on a subset of the training set and a subset of the test set. Also, we occasionally print out samples from the model.
- 3-D visualization during model training. It's fundementally
important, I think, to visualize the 3-D structure of proteins
output by the language model during training. To do this, I use
ESMFold (either via
curlor local install). ESMFold will rapidly (a second or two) fold small proteins like those in the AcyP example dataset so you can easily keep track of the folding progress as the model trains
To run the code locally on Metal, simply pass --device mps to the main script.
To install the dependencies (just PyTorch and Biotite, optionally Hugging Face
tokenizers if needed)
# virtual environment
python -m venv .venv
python -m pip install torch rich biotite plotly tensorboard
To instantiate a new model and train on a FASTA dataset, use the following command line. The example dataset in the repo contains 26,878 homologs of AcyP that are between 64 and 128 residues in length and contain predicted catalytic residue sites from UniProt.
To train a model on this dataset, use the following command:
python main.py -i acyp.fa -o acyp
Once the model is trained, and you would like to sample (data will be printed in FASTA format):
python main.py -i acyp.fa -o acyp --sample-only
Let's examine how different levels of input clustering change the performance of a small transformer model. To do this, let's create a small model and then train it on the same dataset, clustered to different levels of sequence identity with MMseqs2.
We'll start with a dataset of AcyP homologs that I collected from UniProt. See the preprocessing notebook for details.
Let's cluster the dataset at three different levels: at 100%, at 90%, and at 50% sequence identity. This reduces our number of samples
# create an mmseqs db
mmseqs createdb acyp.fa DB
# cluster to 100% identity
mmseqs cluster -c 1.00 --min-seq-id 1.00 DB clust100 tmp
mmseqs createsubdb clust100 DB rep100
mmseqs convert2fasta rep100 rep100.fa
# 90% identity
mmseqs cluster -c 0.95 --min-seq-id 0.90 DB clust90 tmp
mmseqs createsubdb clust90 DB rep90
mmseqs convert2fasta rep90 rep90.fa
# 50% identity
mmseqs cluster -c 0.80 --min-seq-id 0.50 DB clust50 tmp
mmseqs createsubdb clust50 DB rep50
mmseqs convert2fasta rep50 rep50.fa This provides the following datasets for us to try:
| Clustering level | Number of sequences |
|---|---|
| No clustering | 26,856 |
| 100% identity | 23,274 |
| 90% | 18,716 |
| 50% | 12,383 |
To establish train/test splits, we'll use the random seed 42, and take 10% of the data at random. At the 90% identity level, that means that, for each protein in the test set, the closest protein in the training set is 90% identical, at maximum.
We'll use a small transformer model, with an embedding size of 64, with 4 layers and 4 heads, and we'll train for 75k steps. Each step will be a minibatch of size 32, with a block size of 128 sampled randomly from the training set.
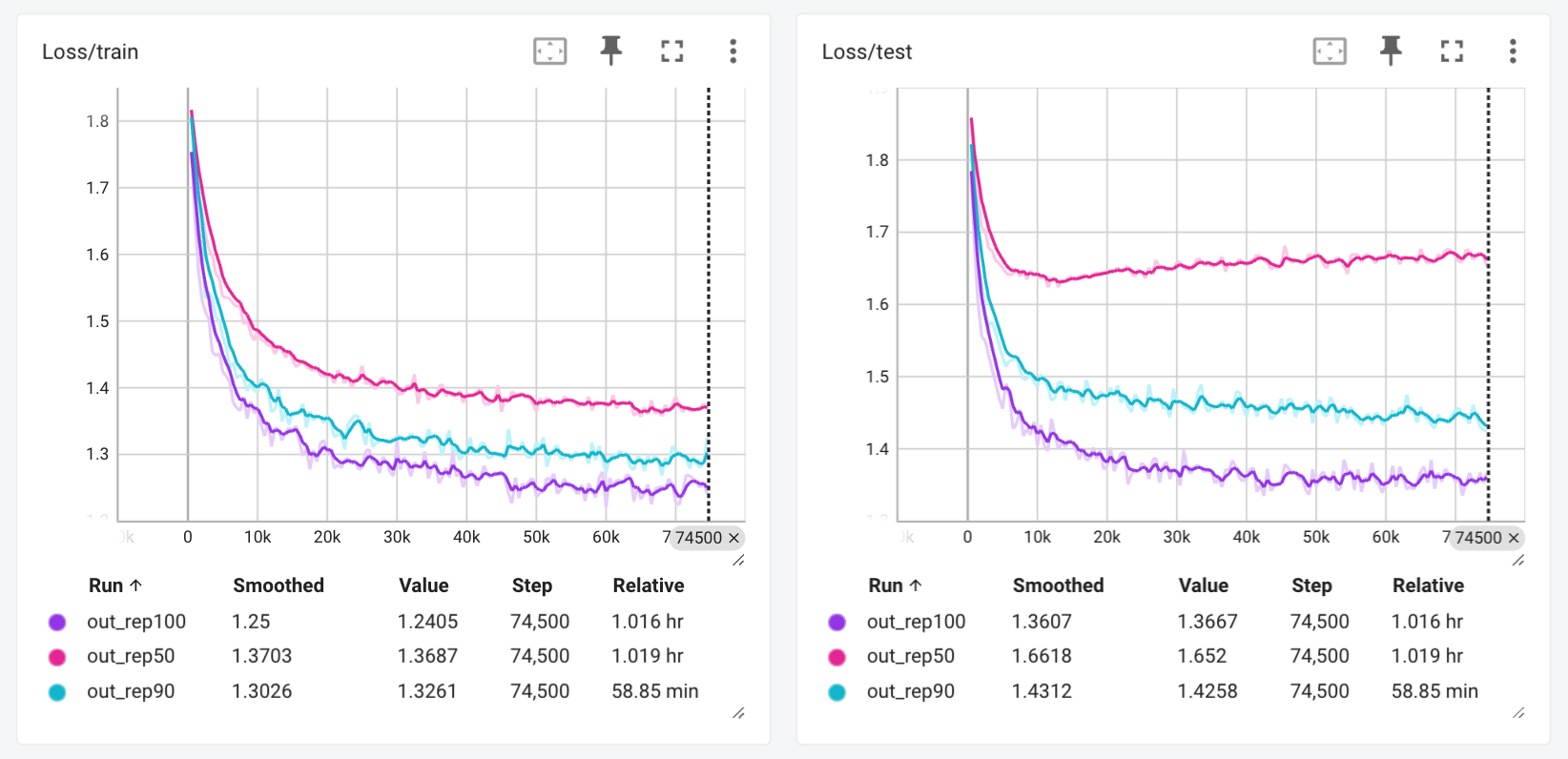
Here is what the training and test loss looks like for the three clusters 100%, 90%, and 50%, just a screenshot of the Tensorboard instance.
This brings me to an important point, which is the importance of clustering your data when working with protein sequences. Here, we can see that while the train loss looks fine for each of the three training runs, at the 50% level, while the training loss goes to under 1.4, the test loss never goes below 1.6, and according to the test loss, the model has stopped learning around step 10k.
The correct level of clustering is an important hyperparameter when training sequence-based models such as this decoder-only transformer model trained on the AcyP dataset.
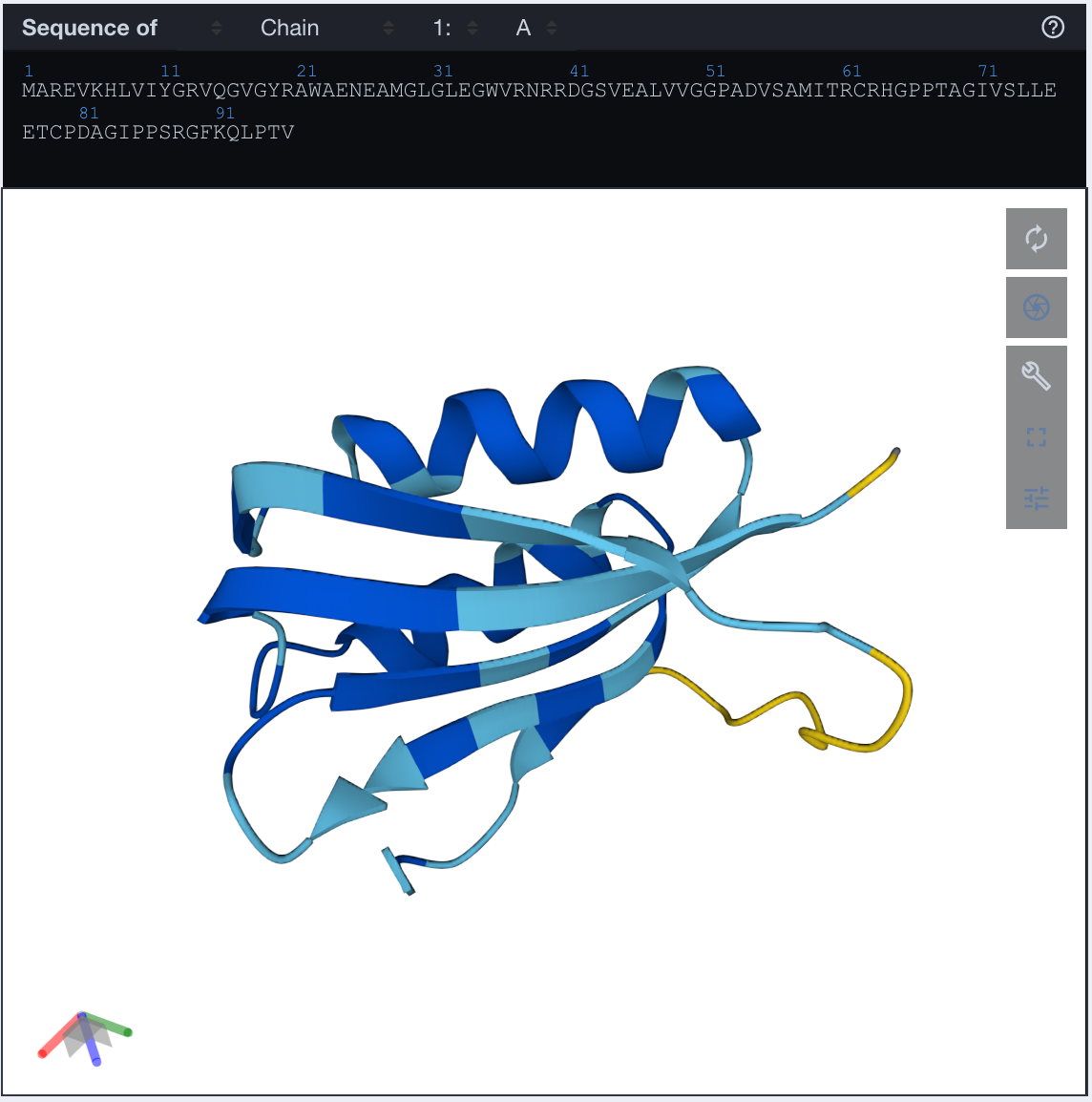
Just for fun, let's generate a couple of samples and fold them. I picked this sample from the last checkpoint
>sample
MAREVKHLVIYGRVQGVGYRAWAENEAMGLGLEGWVRNRRDGSVEALVVGGPADVSAMITRCRHGPPTAGIVSLLEETCPDAGIPPSRGFKQLPTV
and folded it with ESMFold, and obtained this structure
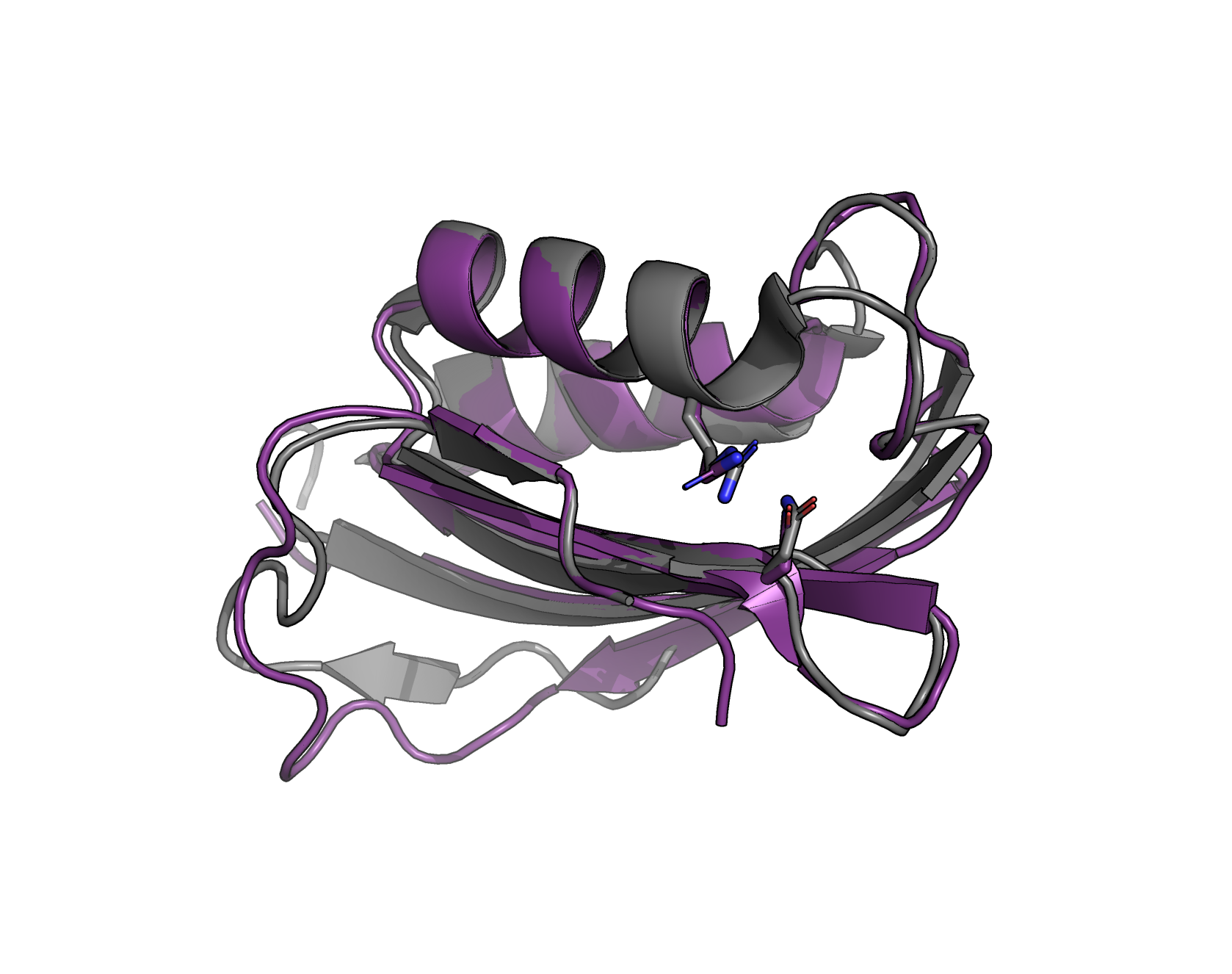
I then was interested to compare this to a crystal structure of a homolog from the family. The crystal structure PDB code 1URR is "novel Drosophila melanogaster acylphosphatase" (Zuccotti+ 2004).
I found that the generated protein is only about 25% identical, at a primary sequence level, to the sequence from 1URR. However, after structural alignment in PyMOL, we can see that not only is the generated sequence predicted to fold into the same overall fold as the real sequence from 1URR, but also both catalytic residues are present and the correct identity (in the crystal, these are R27 and N45).
Superposition of generated protein and crystal structure. The crystal, in gray, is a acylphosphatase from Drosophila melanogaster with PDB code 1URR. The generated protein, as folded by ESMFold, is depicted purple. Figure drawn with PyMOL.