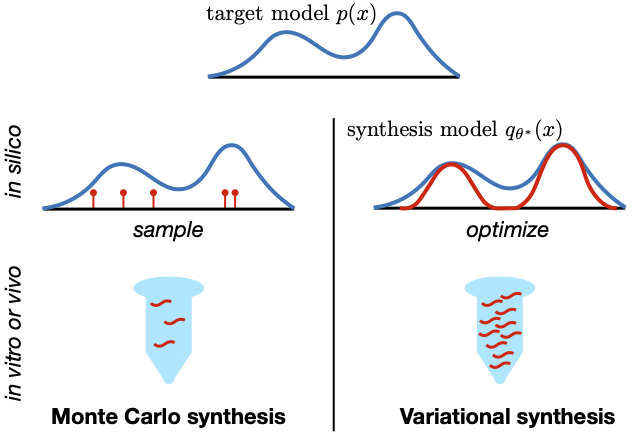
This repo provides example code implementing variational synthesis, as introduced in the paper
Optimal Design of Stochastic DNA Synthesis Protocols based on Generative Sequence Models. Eli N. Weinstein, Alan N. Amin, Will Grathwohl, Daniel Kassler, Jean Disset, Debora S. Marks. AISTATS. 2022.
Download this repo, create a new virtual environment (e.g. using conda) with python 3.9, and run
pip install .
To test your installation, navigate to the VariationalSynthesis subfolder and run
pytest
To optimize an example stochastic synthesis model, navigate to the VariationalSynthesis subfolder and run
python model.py data/config_ex.cfg
A detailed description of the method's options can be found in the config file data/config_ex.cfg.
Data: Stochastic synthesis models are optimized on samples from target distributions: this can be controlled with the [general][target_samples] option in the config file. In particular, the datasets used in the paper are samples from a DHFR Potts model (which can be found in data/DHFR_Potts.fasta), raw evolutionary DHFR sequences (data/DHFR_raw.fasta), samples from a GFP ICA-MuE model (data/GFP.fasta) and a TCR ICA-MuE model (data/TCR.fasta).
Models: Our synthesis models use either fixed or combinatorial assembly: this can be controlled with the [model][assembly] option in the config file. Our synthesis models also use either enzymatic mutagenesis, finite nucleotide mixtures, finite codon mixtures or arbitrary codon mixtures: this can be controlled with the [model][unit] and [model][constraint] options in the config file.
Results: Results will (by default) be put in a time-stamped subfolder within the data/logs folder. The example input (data/config_ex.cfg) trains a finite codon mixture model with fixed assembly and M=10 on the raw DHFR dataset. In the output config file (the [results][perpl_per_res] section), we report the perplexity of the trained synthesis model: 9.2, which matches Figure 3D of the paper. The trained parameters of the model are pickled in the output params.pkl file; they can be loaded from within python with
import pickle
from VariationalSynthesis.model import SynthesisModel
with open('data/logs/TIMESTAMP/params.pkl', 'rb') as pr:
model = pickle.load(pr)
with TIMESTAMP replaced by the time-stamp of your results.