Giorgia Graells and Derek Corcoran 2022-01-09
This repository serves to document and store the analyses and results for the manuscript Exploring habitat use of terrestrial and marine birds in urban coastal areas sent to the jounrnal Frontiers Ecology And Evolution
First we load the required packages
# For manipulating and reading raster datasets
library(raster)
library(terra)
# For cleaning datasets
library(tidyverse)
# For managing vector spatial datasets
library(sf)
# For caclulating occupancy models
library(unmarked)
# For selecting models
library(MuMIn)Then we load the coordinates of the sampling sites for the surveys and
transform them in to a SpatVector object:
Puntos_Hull <- read_csv("https://raw.github.com/derek-corcoran-barrios/LayerCreationBuffer/main/Coords.csv") %>%
mutate(geometry = str_remove_all(str_remove_all(str_remove_all(geometry, "c"), "\\("), "\\)"))
Puntos_Hull$Lon <- str_split(Puntos_Hull$geometry, pattern = ",", simplify = T)[,1] %>% as.numeric()
Puntos_Hull$Lat <- str_split(Puntos_Hull$geometry, pattern = ", ", simplify = T)[,2] %>% as.numeric()
Puntos_Hull <- Puntos_Hull %>%
dplyr::select(-geometry) %>%
st_as_sf(coords = c(2,3), crs = 4326) %>%
st_transform(crs = "+proj=utm +zone=19 +south +datum=WGS84 +units=m +no_defs") %>%
terra::vect()Then we generate a vector of the distances used to calculate the proportion of landuse in meters as seen in Graells and Corcoran (2022):
Distancias <- round(seq(from = 30, to = 5000, length.out = 10), -2)
Distancias[1] <- 30We then download the rasters from that repository to generate the
Layers list with one raster stack for each distance, and another list
called OccuVars where we extract the values for the proportion of each
landuse for each one of the points
Layers <- list()
OccuVars <- list()
for(i in 1:length(Distancias)){
Layers[[i]] <- terra::rast(paste0("/vsicurl/https://raw.github.com/derek-corcoran-barrios/LayerCreationBuffer/main/Proportions_", Distancias[i],".tif"))
OccuVars[[i]] <- terra::extract(Layers[[i]], Puntos_Hull)
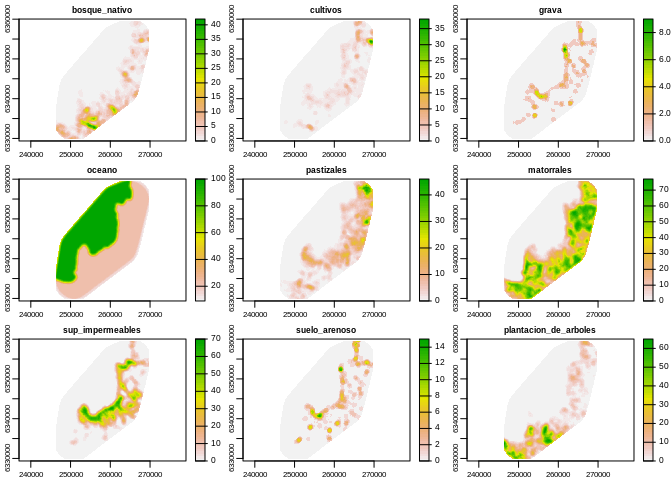
}As an example in 1.1, we can see the proportions of each type of landuse for each point in the study site 600 meters around them.
Just as another example in Table 1.1 we see the extracted values for the first 10 sites of the study:
| ID | bosque_nativo | cultivos | grava | oceano | pastizales | matorrales | sup_impermeables | suelo_arenoso | plantacion_de_arboles |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 0 | 0 | 0 | 76 | 2 | 7 | 9 | 1 | 1 |
| 2 | 1 | 1 | 1 | 55 | 4 | 14 | 21 | 1 | 2 |
| 3 | 1 | 2 | 1 | 54 | 5 | 23 | 8 | 1 | 3 |
| 4 | 1 | 1 | 1 | 52 | 4 | 16 | 20 | 1 | 3 |
| 5 | 1 | 1 | 0 | 51 | 5 | 20 | 18 | 1 | 2 |
| 6 | 5 | 0 | 0 | 33 | 2 | 32 | 1 | 0 | 15 |
| 7 | 1 | 1 | 1 | 55 | 3 | 22 | 13 | 1 | 2 |
| 8 | 1 | 1 | 1 | 57 | 4 | 22 | 8 | 1 | 3 |
| 9 | 1 | 1 | 1 | 60 | 3 | 23 | 9 | 1 | 2 |
| 10 | 1 | 2 | 1 | 47 | 6 | 25 | 8 | 1 | 3 |
Table 1.1: The values of the proportion for the first ten sites of the study
We used the function batchoccu2 which is a modification of the
batchoccu2, from the DiversityOccupancy package (Corcoran et al.
2017). This function first fits all possible combinations of the
probability of detection of a species, and selects the best model by
AICc, and then using that model, for probability of detection, it tests
all possible models for occupancy given that model and selects the best
by AICc. In the next code
batchoccu2 <- function(pres, sitecov, obscov, spp, form, SppNames = NULL, dredge = FALSE) {
if(is.null(SppNames)){
SppNames <- paste("species", 1:spp, sep =".")
}
secuencia <- c(1:spp)*(ncol(pres)/spp)
secuencia2<-secuencia-(secuencia[1]-1)
models <- vector('list', spp)
fit <- matrix(NA, nrow(pres), spp)
Mods <- list()
if(is.null(SppNames)){
colnames(fit) <- paste("species", 1:spp, sep =".")
}else if(class(SppNames) == "character"){
colnames(fit) <- SppNames
}
if (dredge == FALSE) {
for(i in 1:length(secuencia)) {
data <- pres[, secuencia2[i]:secuencia[i]]
data2 <- unmarkedFrameOccu(y = data, siteCovs = sitecov, obsCovs = obscov)
try({
models[[i]] <- occu(as.formula(form), data2)
}, silent = T)
try({
fit[, i] <- suppressWarnings(predict(models[[i]], type = "state", newdata = sitecov))$Predicted
}, silent = T)
Mods = NULL
print(paste("Species", as.character(i), "ready!"))
}
}
else {
for(i in 1:length(secuencia)) {
data <- pres[, secuencia2[i]:secuencia[i]]
data2 <- unmarkedFrameOccu(y = data, siteCovs = sitecov, obsCovs = obscov)
try({
#Partimos en dos Detección y occupancia
form <- as.character(form)
Div <- str_squish(form) %>% str_remove_all(" ") %>% stringr::str_split(pattern = "~", simplify = T)
### Separamos dos formulas Occupancia y Deteccion
Det <- Div[length(Div) - 1]
VarDet <- str_split(Det, "\\+", simplify = T) %>% as.character()
Fs <- list()
print(paste("Starting to fit detection models for species", i, "of", length(secuencia)))
for(x in 1:(length(VarDet) + 1)){
if(x == (length(VarDet) + 1)){
Formulas <- data.frame(Form = "~1 ~ 1", AICc = NA)
Formulas$AICc[j] <- try(MuMIn::AICc(occu(as.formula("~1 ~1"), data2)), silent = T)
}else{
Test <- combn(VarDet, x, simplify = F)
Formulas <- data.frame(Form = rep(NA, length(Test)), AICc = rep(NA, length(Test)))
for(j in 1:length(Test)){
Temp <- paste("~", paste(Test[[j]], collapse = " + "), "~ 1")
Formulas$Form[j] <- Temp
Temp <- as.formula(Temp)
Formulas$AICc[j] <- try(MuMIn::AICc(occu(Temp, data2)), silent = T)
gc()
}
}
Fs[[x]] <- suppressWarnings(Formulas %>% mutate(AICc = as.numeric(AICc)) %>% dplyr::filter(!is.na(AICc)) %>% arrange(AICc))
message(paste("finished for", x, "number of variables"))
}
Fs <- suppressWarnings(purrr::reduce(Fs, bind_rows) %>% arrange(AICc))
Selected <- Fs$Form[1] %>% str_split("~", simplify = T) %>% as.character()
Selected <- Selected[length(Selected) - 1] %>% str_squish()
print(paste("Detection model for species", i, "is", Selected))
Occup <- Div[length(Div)]
VarOccup <- str_split(Occup, "\\+", simplify = T) %>% as.character()
Fs <- list()
print(paste("Starting to fit occupancy models for species", i, "of", length(secuencia)))
for(x in 1:(length(VarOccup) + 1)){
if(x == (length(VarOccup) + 1)){
Formulas <- data.frame(Form = paste("~",Selected, "~ 1"), AICc = NA)
Formulas$AICc[j] <- try(MuMIn::AICc(occu(as.formula(paste("~",Selected, "~ 1")), data2)), silent = T)
}else{
Test <- combn(VarOccup, x, simplify = F)
Formulas <- data.frame(Form = rep(NA, length(Test)), AICc = rep(NA, length(Test)))
for(j in 1:length(Test)){
Temp <- paste("~", Selected, "~", paste(Test[[j]], collapse = " + "))
Formulas$Form[j] <- Temp
Temp <- as.formula(Temp)
Formulas$AICc[j] <- try(MuMIn::AICc(occu(Temp, data2)), silent = T)
if((j %% 100) == 0){
message(paste(j, "of", length(Test), "Ready"))
gc()
}
}
}
Fs[[x]] <- suppressWarnings(Formulas %>% mutate(AICc = as.numeric(AICc)) %>% dplyr::filter(!is.na(AICc)) %>% arrange(AICc))
message(paste("finished for", x, "number of variables", Sys.time()))
}
Fs <- suppressWarnings(purrr::reduce(Fs, bind_rows) %>% arrange(AICc))
Mods[[i]] <- Fs
Best <- Fs$Form[1]
models[[i]] <- occu(as.formula(Best), data2)
#dredged <- suppressWarnings(dredge(occu(form, data2)))
# select the first model and evaluate
#models[[i]] <- eval(getCall(dredged, 1))
}, silent = T)
try({
#predictions for the best model
fit[, i] <- suppressWarnings(predict(models[[i]], type = "state", newdata = sitecov))$Predicted
}, silent = T)
print(paste("Species", as.character(i), "ready!"))
}
}
if(is.null(SppNames)){
names(models) <- paste("species", 1:spp, sep =".")
}else if(class(SppNames) == "character"){
names(models) <- SppNames
}
if(is.null(SppNames)){
names(Mods) <- paste("species", 1:spp, sep =".")
}else if(class(SppNames) == "character" & !is.null(Mods)){
names(Mods) <- SppNames
}
cond <- sapply(models, function(x) !is.null(x))
models <- models[cond]
fit <- fit[,cond]
Not <- SppNames[!(cond)]
if(sum(!cond) >= 1){
message(paste("species", paste(Not, collapse = ", "), "did not converge, try with less variables"))
}
result <- list(Covs = sitecov, models = models, fit = fit, Mods = Mods)
class(result)<- "batchoccupancy"
return(result)
}Corcoran, Derek, Dylan Kesler, Lisa Webb, and Giorgia Graells. 2017. DiversityOccupancy: Building Diversity Models from Multiple Species Occupancy Models. https://CRAN.R-project.org/package=DiversityOccupancy.
Graells, Giorgia, and Derek Corcoran. 2022. Genearation of layers of proportions of landuse at different distances for the Valparaíso, Viña del Mar and Concón communes in Chile (version 0.0.1). https://github.com/derek-corcoran-barrios/LayerCreationBuffer.