The TinDaisy-Core pipeline is a modular software package designed for detection of somatic variants from tumor and normal exome data. TinDaisy-Core obtains variant calls from four callers, merges them, and applies various filters.
Callers used:
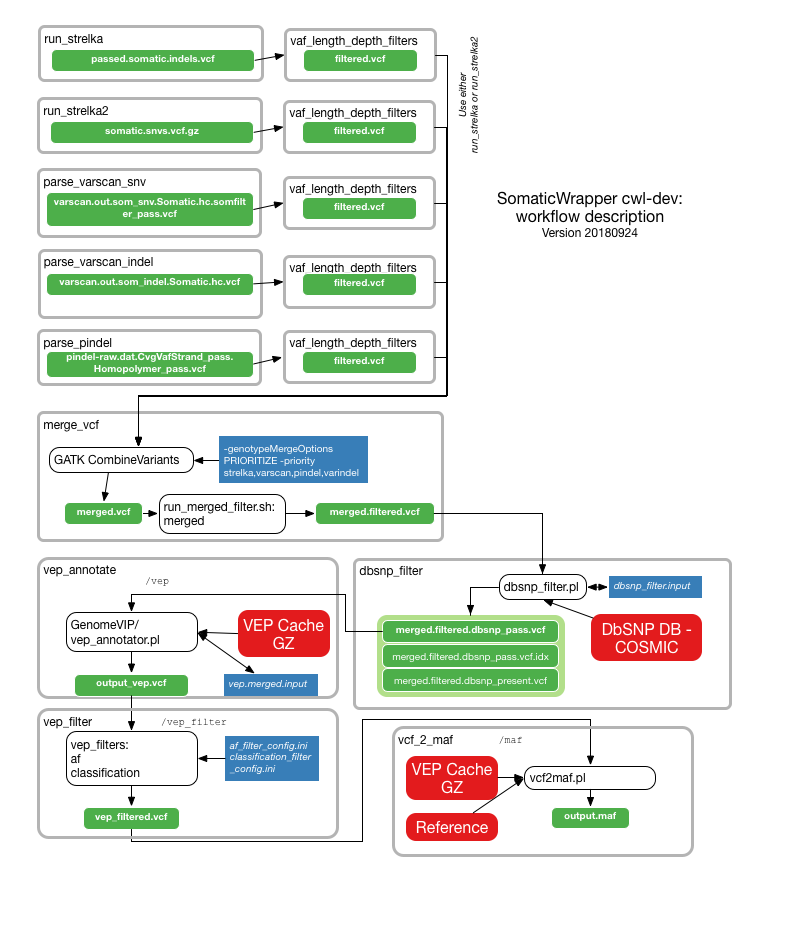
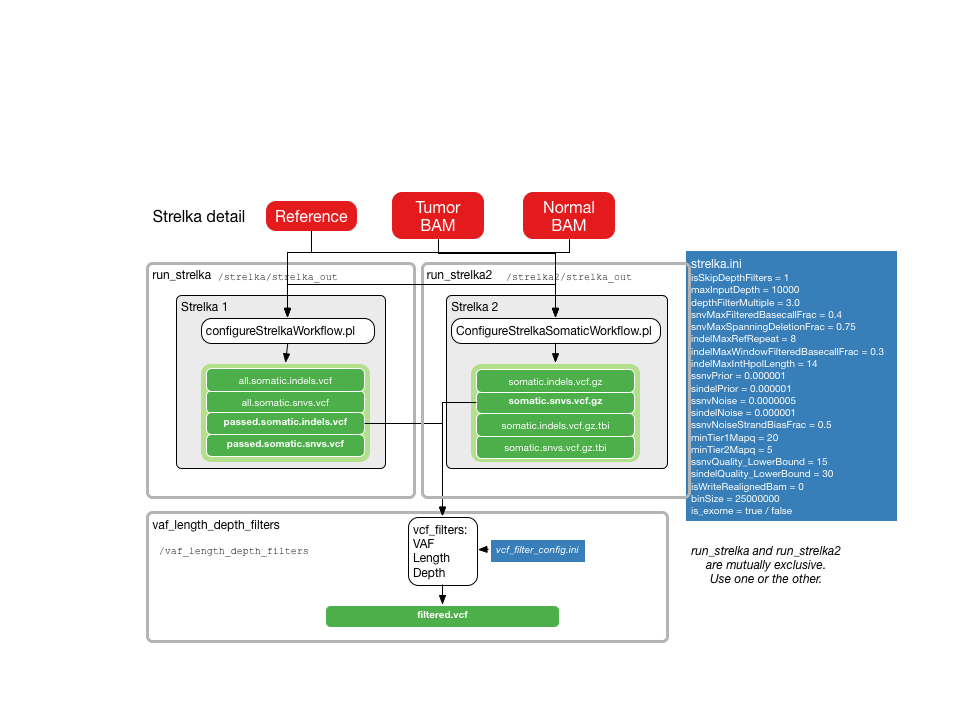
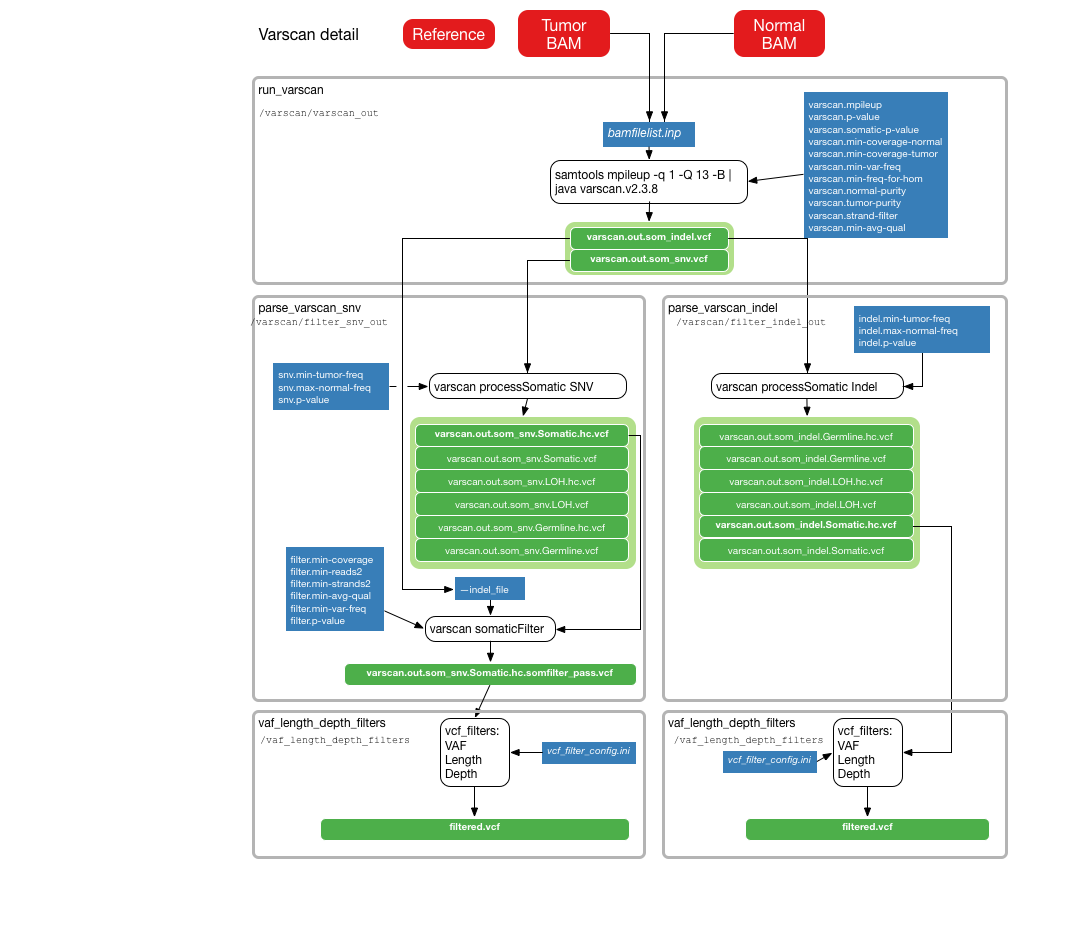
SNV calls from Strelka2, Varscan, Mutect. Indel calls from Stralka2, Varscan, and Pindel. CWL Mutect Tool is used for CWL Mutect calls
Filters applied (details in VCF output)
- For indels, require length < 100
- Require normal VAF <= 0.020000, tumor VAF >= 0.050000 for all variants
- Require read depth in tumor > 14 and normal > 8 for all variants
- All variants must be called by 2 or more callers
- Require Allele Frequency < 0.005000 (as determined by vep)
- Retain exonic calls
- Exclude calls which are in dbSnP but not in COSMIC
TinDaisy and TinDaisy-Core were developed from SomaticWrapper and GenomeVIP.
See TinDaisy for details about installation and usage of TinDaisy-Core in a CWL environment
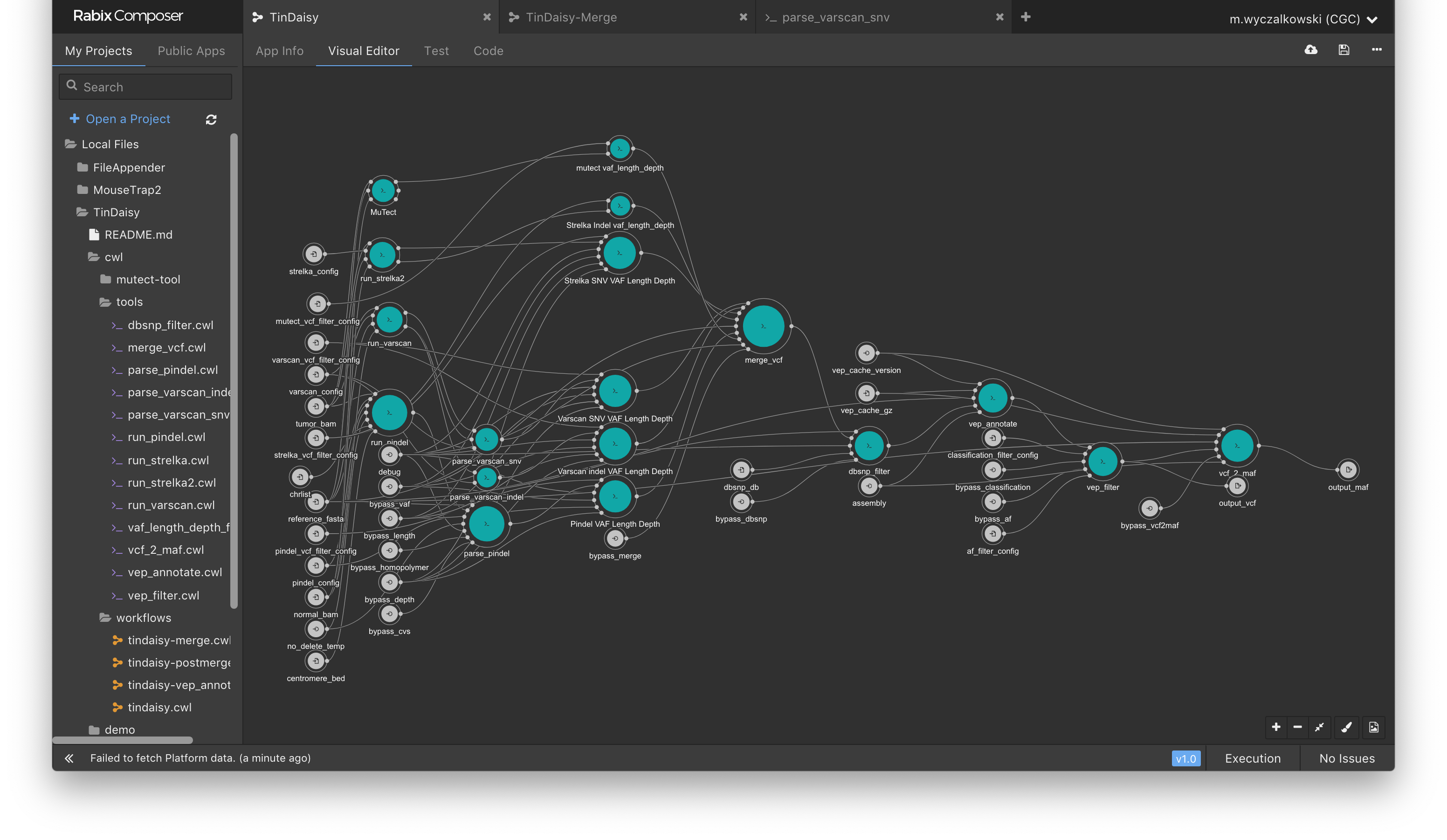
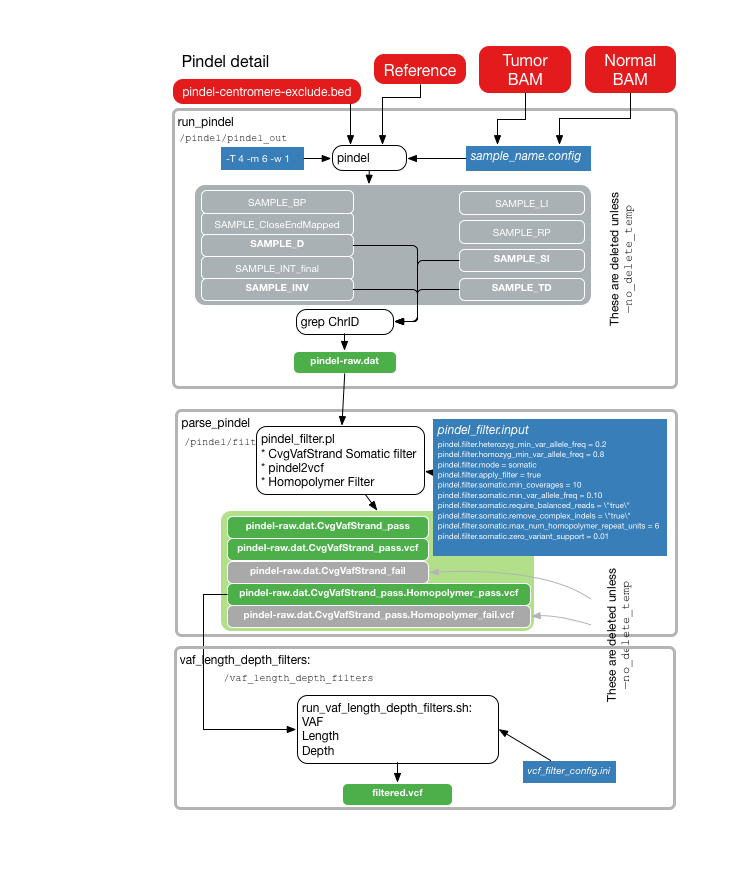
Below are visual outlines of the TinDaisy-Core workflow. Note that the actual workflow is implemented at CWL level in TinDaisy.
The current CWL implementation is visualized below using Rabix Composer
The following illustrations provide details about the internals of each TinDaisy-Core step. Note that the Overall figure is outdated, as it does not incorporate Mutect SNV calls.
- Matthew Wyczalkowski m.wyczalkowski@wustl.edu
- Song Cao scao@wustl.edu
- Jay Mashl rmashl@wustl.edu