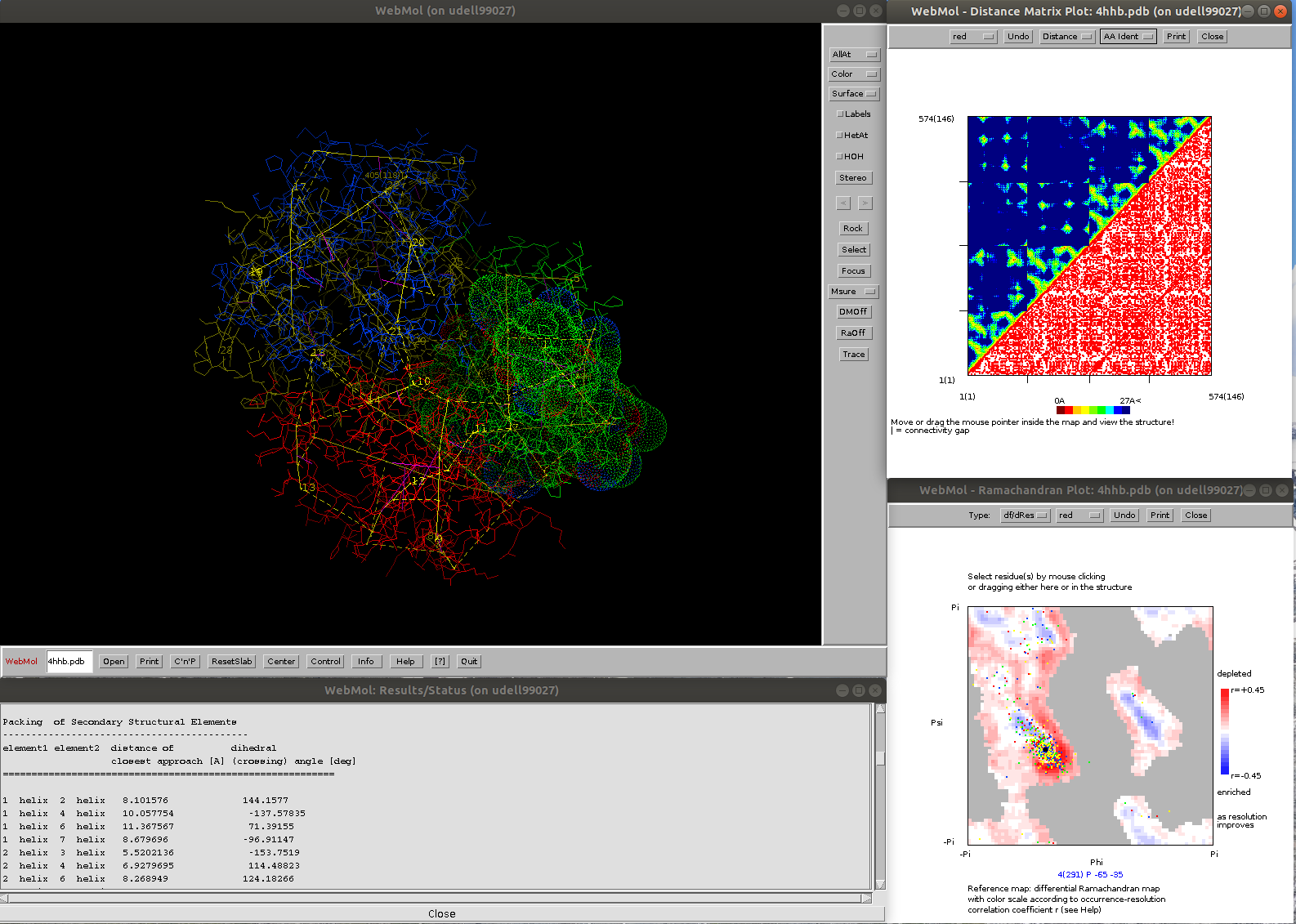
Java-based protein structure viewer and analysis program
Launch WebMol (Windows: double click on jar file, Linux: java -jar webmol.jar) Load structure in PDB-file format, either by invoking "Open" or, under Linux, as a parameter: java -jar webmol.jar
Example: java -jar webmol.jar https://files.rcsb.org/view/4HHB.pdb
For help/instructions, see "Help"
Reference: Walther, D (1997) WebMol--a Java-based PDB viewer. Trends in biochemical sciences 22 (7), 274-275
Source code available upon request. WebMol was developed at the EMBL (Heidelberg, Germany) and UCSF (San Francisoco, USA) in the years 1996-1999
Author: Dirk Walther walther@mpimp-golm.mpg.de
Unique features, highlights
- Analysis of packing geometries of secondary structural elements (axes, distances, and angles)
- surface and cavity computations, including surface and volume area computation
- interactive distance matrix and Ramachandran plots, with differential Ramachandran map as reference (see https://pubmed.ncbi.nlm.nih.gov/10089363/)
- API available (see below)
java.lang.Object
|
+----java.awt.Component
|
+----java.awt.Container
|
+----java.awt.Window
|
+----java.awt.Frame
|
+----webmol
- public class webmol
- extends Frame
WebMol - Java PDB structure viewing and analysis program Author: Dirk Walther (walther@mpimp-golm.mpg.de) Version March 10, 1998European Molecular Biology Laboratory (EMBL) June 1996-March 1997 University of California, San Francisco (UCSF) April 1997-1999
Reference: Walther D. (1997) WebMol - a Java based PDB viewer. Trends Biochem Sci, 22: 274-275
Example ("webmolExample.java")
import webmol;
public class webmolExample
{
public static void main(String args[])
{
int num,add=1,col;
webmol wm=new webmol("pdb1ppt.ent",700,600);
// webmol wm=new webmol("http://www.embl-heidelberg.de/~walther/JAVA/pdb/1ppt.brk",700,600);
// reads it across the web
// webmol wm=new webmol("https://files.rcsb.org/view/1PPT.pdb",700,600);
// reads it from PDB directly
num=wm.getNumberOfResidues();
int res=0;
for(int j=0;j<2*num;j++)
{
if(wm.getResidueType(res)=='P') // if amino acid = proline then color yellow
col=wm.yellow;
else
col=wm.current; // current color set by WebMol
wm.select(res,
wm.SIDECHAIN_VS_SIDECHAIN,
col,
wm.THICK);
wm.redraw(500);
if(res==num-1) add=-1;
if(res==0) add=1;
wm.unselect(res);
res+=add;
}
wm.close(true);
}
}
-
blue
- Color to apply for selection highlighting
- current
- Color to apply for selection highlighting
- cyan
- Color to apply for selection highlighting
- gray
- Color to apply for selection highlighting
- lilac
- Color to apply for selection highlighting
- orange
- Color to apply for selection highlighting
- pink
- Color to apply for selection highlighting
- red
- Color to apply for selection highlighting
- yellow
- Color to apply for selection highlighting
- SELECTED_ONLY
- Select method
- SIDECHAIN_VS_BACKBONE
- Select method
- SIDECHAIN_VS_SIDECHAIN
- Select method
- THICK
- thick lines
- THIN
- thin lines
- webmol(String)
- launching WebMol with default width and height, upon closing the WebMol frame all Java programs will be exited.
- webmol(String, boolean)
- webmol(String, boolean, int, int)
- webmol(String, int, int)
- close()
- close WebMol
- close(boolean)
- close Webmol and exit (exit=true)/ not exit (exit=false) all current Java programs under which WebMol was running
- getChainID(int)
- returns the chain identifier of residue res
- getNumberOfResidues()
- returns the number of amino acid residues in the current protein structure
- getPDBSeqNum(int)
- returns the PDB sequence number of residue res as it may differ from the consecutive enumeration used by WebMol
- getResidueType(int)
- returns the amino acid type (one-letter code) of residue res
- redraw()
- redraw the structure in the WebMol main window
- redraw(int)
- redraw the structure in the WebMol main window
- select(int, int, int, int)
- select a single residue, enumeration of residues is in consecutive order; i.e.
- unselect(int)
- unselect residue
- unselectAll()
- undo all selections
public static final int THICK
- thick lines
public static final int THIN
- thin lines
public static final int current
- Color to apply for selection highlighting
public static final int red
- Color to apply for selection highlighting
public static final int blue
- Color to apply for selection highlighting
public static final int yellow
- Color to apply for selection highlighting
public static final int lilac
- Color to apply for selection highlighting
public static final int gray
- Color to apply for selection highlighting
public static final int pink
- Color to apply for selection highlighting
public static final int cyan
- Color to apply for selection highlighting
public static final int orange
- Color to apply for selection highlighting
public static final int SELECTED_ONLY
- Select method
public static final int SIDECHAIN_VS_BACKBONE
- Select method
public static final int SIDECHAIN_VS_SIDECHAIN
- Select method
public webmol(String Protein,
boolean invoked,
int width,
int height)
- Parameters:
- Protein - PDB-file
- invoked - determines how to respond to closures of the WebMol Frame; true: close WebMol only, false: close WebMol and all Java programs under which WebMol was running
- width - frame width
- height - frame height
public webmol(String Protein,
int width,
int height)
- Parameters:
- Protein - PDB-file
- width - width of the WebMol frame
- height - height of the WebMol frame
public webmol(String Protein)
- launching WebMol with default width and height, upon closing the WebMol frame all Java programs
will be exited.
- Parameters:
- Protein - PDB-file
public webmol(String Protein,
boolean invoked)
- Parameters:
- Protein - PDB-file
- invoked - set true to exit WebMol without closing the program that invoked WebMol
public void close()
- close WebMol
public void close(boolean exit)
- close Webmol and exit (exit=true)/ not exit (exit=false) all current Java programs under which WebMol was running
- Parameters:
- exit - true for exit all programs, false for just leaving WebMol
public void select(int res,
int selectMethod,
int color,
int thick)
- select a single residue, enumeration of residues is in consecutive order;
i.e. from 0 to num-1, irrespective of pdb enumerations
- Parameters:
- res - residue number (consecutive order
- selectMethod - method of highlighting selected residues, values can be SELECTED_ONLY (display selected residues only), SIDECHAIN_VS_BACKBONE (display sidechains of selected residues against the protein backbone), SIDECHAIN_VS_SIDECHAIN (keep displaying all residues against highlighted selected residues)
- color - color to be used for highlighting selected residue (see Fields)
- thick - line thickness (THICK or THIN)
public void unselect(int res)
- unselect residue
- Parameters:
- res - residue number
public void unselectAll()
- undo all selections
public int getNumberOfResidues()
- returns the number of amino acid residues in the current protein structure
public int getPDBSeqNum(int res)
- returns the PDB sequence number of residue res as it may differ
from the consecutive enumeration used by WebMol
- Parameters:
- res - residue (consecutive order)
public char getResidueType(int res)
- returns the amino acid type (one-letter code) of residue res
- Parameters:
- res - residue (consecutive order)
public char getChainID(int res)
- returns the chain identifier of residue res
- Parameters:
- res - residue (consecutive order)
public void redraw(int delay)
- redraw the structure in the WebMol main window
- Parameters:
- delay - redraw after milliseconds
public void redraw()
- redraw the structure in the WebMol main window