Automated on-the-fly fiducial-based tilt-series alignment for Warp tilt-series directories
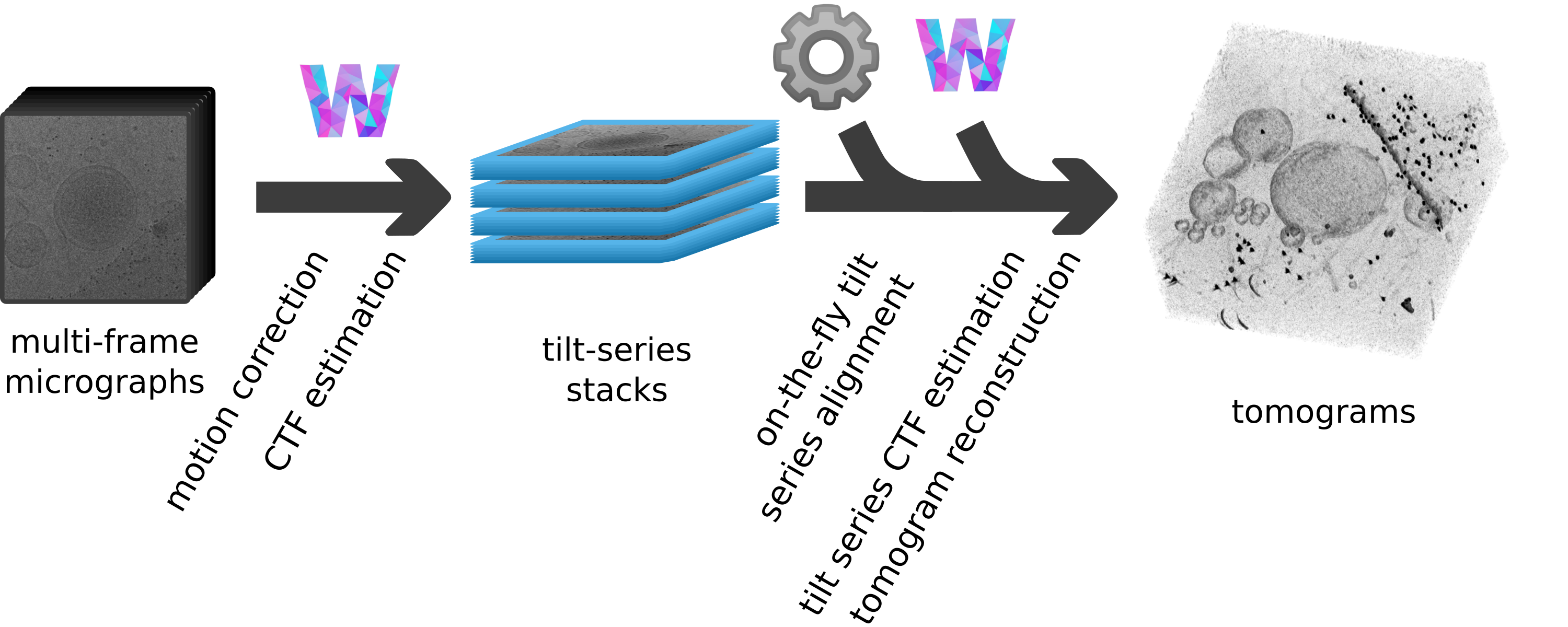
Workflow from raw data to aligned tilt-series
- Follow the Warp user guide to take you from frames to tilt-series
- Run
dautoalign4warp, it will watch your tilt-series directory (imod) and align any incoming tilt-series - Import the results into warp and reconstruct your tomograms!
- (optional) Use
warp2catalogueto easily create a Dynamo catalogue for particle picking
Motivation
- Robust, automated tilt-series alignment of stacks created by Warp
- Ability to easily use Warp reconstructions with the geometrical particle picking tools in Dynamo
Outline
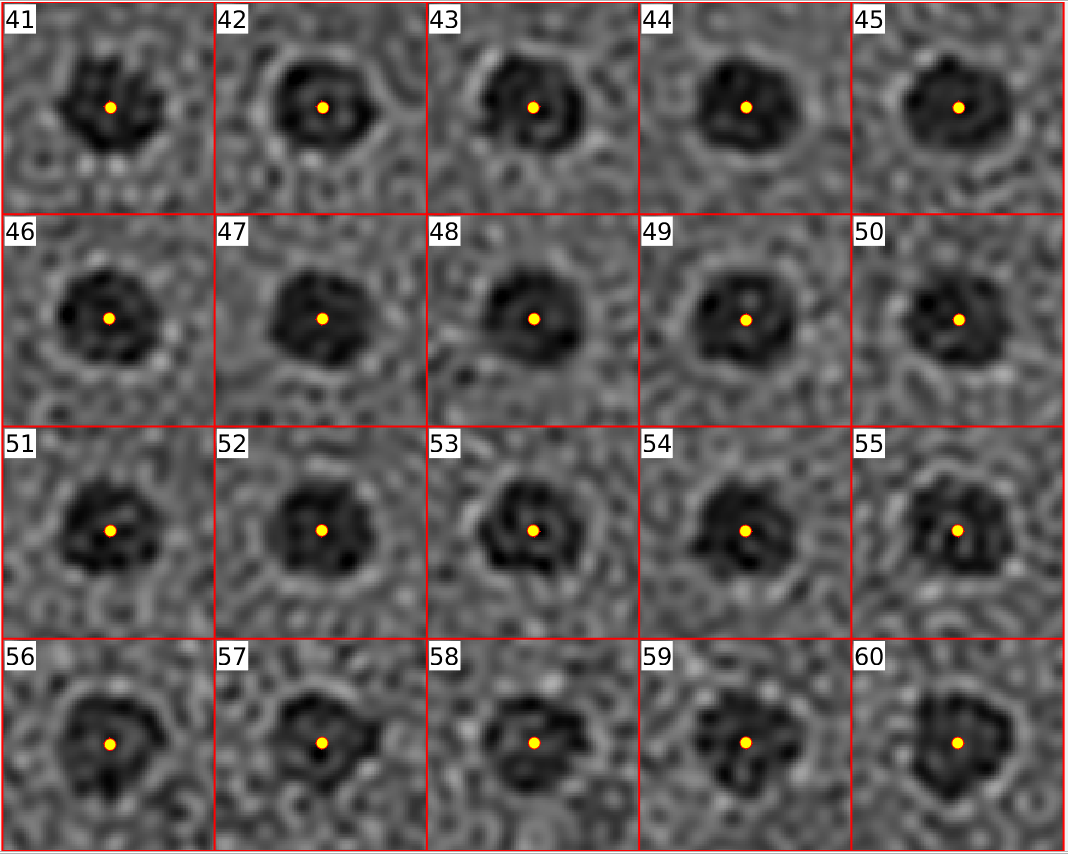
- Automated robust fiducial detection and fiducial position refinement using Dynamo
- Creation of an IMOD model from these positions
- Solve for tilt-series alignment transformations using the IMOD program tiltalign (fixed tilt angles, solve for one rotation angle, robust fitting)
- Preparation of all results for easy import back into Warp
Particle picking in Dynamo?
Do you want to use the geometrical tools in Dynamo to pick particles, imparting prior knowledge about your particle orientation to further subtomogram averaging?
If so, you may want to use the warp2catalogue function to quickly set up a catalogue of your data, correctly linked for visualisation of Warp's deconvolved tomograms with cropping directly from the 3d-CTF corrected tomogram from warp
Installation
Requirements
- Dynamo (1.1.478 or later, 1.1.522 has problems) activated in MATLAB
- IMOD (tested on 4.10.35)
- MATLAB (R2019a or later) (tested with the following toolboxes)
- parallel computing toolbox
- image processing toolbox
- computer vision toolbox
- curve fitting toolbox
- Python (3.0 or later)
There have been reports of slowness when MATLAB is configured to run on external computational resources, for this reason we recommend to run locally.
Download and install
git clone https://github.com/alisterburt/autoalign_dynamo.git
cd autoalign_dynamo
./install.shThis requires git, which can be installed by your package manager if not already present in your system, for example...
sudo apt-get install gitYou will be asked where you would like to install the package, the default location is...
/opt/autoalign_dynamoAfter installation you can remove the installation files
cd ..
rm -r autoalign_dynamoActivation and Running
- Make sure Dynamo is activated in MATLAB (
dynamo_activate) - Activate autoalign_dynamo (
run /path/to/install/autoalign_activate.m) - run
dautoalign4warp(<warp_tilt_series_directory>, <pixel_size_angstrom>, <fiducial_diameter_nm>, <nominal_rotation_angle>, <output_folder>)
This will watch the warp tilt-series directory, align your tilt-series on-the-fly and tidy everything up ready for import back into Warp so you can generate your tomograms. The warp tilt-series directory is the 'imod' directory found inside your main warp processing directory.
If you then want to use Dynamo for particle picking...
- Import alignments into Warp as described in Warp's user guide
- Reconstruct downsampled tomograms (and deconvolved tomograms for visualisation) in Warp
- Run
warp2catalogue(<warp_reconstruction_folder>, <pixel_size_angstrom>)
This will generate a catalogue in which the visualisation volume is the deconvolved tomogram from Warp, particles will be cropped from the 3D-CTF corrected, unfiltered reconstructions
To look at this catalogue, in MATLAB type...
dcm warp_catalogue
Additional info
If you want to use the automated alignment script outside of the Warp directory structure, a function called align is provided which allows automated alignment of a single tilt-series.
Enjoy!