This is a set of scripts that try to answer the question: is it possible that a gene A is under control of transcription factor B, given currently available experimental evidence. Response of a gene called Hand2 to progesterone and estrogen was the original topic.
Progesterone is not a library - the scripts are loosely connected by a couple of common methods in the utils module. They average some hundred lines in length, and can probably be figured out without much explanation. They have been tested to run under python 3.6.6, and are supposed to be run in the order in which they are enumerated. They answer a series of sub-questions, which may add up to an answer. Depends on the answer you are hoping for.
- Dependencies
- Organizing work/data flow
- Gene coordinates
- What's with this TAD business
- Which TAD does my gene belong to
- Where do transcription factors bind within that TAD
- How are TF binding sites distributed across TADs
- Where exactly (and and how good) is the TF binding motif within each ChIPSeq region
- Which regions within the TAD come in contact (and how often)
- Change in chromatin accessibility upon progesterone administration?
- Putting it all together
- So, functional TF sites or not?
- TODO
Progesterone pipeline depends on several data sources and python packages. You don't have to install them right away but only (or when) they become necessary.
-
MySQLdb python module, installed with sudo apt install python3-mysqldb. Progesterone scripts also count on 2 mysql configuration files somewhere at the top of the current user's directory tree. One is called '.ucsc_mysql_conf' and looks like this:
[client]
skip-auto-rehash
user = genome
host = genome-mysql.soe.ucsc.edu
port = 3306
The other one is '.mysql_conf' providing credentials for logging into MySQL running locally. You can template it on the one above.
-
data from HiC experiment on ... something - adapt this to the cell/tissue type you are interested in
-
ChIPSeq regions from ENCODE experiment, collected in UCSC genome db
-
CrossMap for transforming coordinates to a single reference assembly (together with transformation chain files, here)
-
Matplotlib python module (pip3 install matplotlib; if your scripts complain about missing tkinter, you might also have to do sudo apt install python3-tk)
-
python tools for handling data in HDF5 format
-
transcription factor (TF) binding motif databases JASPAR and Hocomoco
-
more ChIPSeq data from GEO database (GSE34927, GSE36455, for example. Again YMMV.)
-
maf_parse from PHAST
-
maf_to_fasta.py - for this one to work you will also need pip3 install bx-python
-
Multiple alignment files (mafs) from UCSC for mouse and human - these are voluminous and optional. If you know the chromsome your gene resides on, you can download maf(s) for that chromosome only.
-
For output, the pipeline expects to see a provided (output) directory called raw_data. If raw_data or, sometimes, its subdirectory is nonexistent, each script will prompt you to create it as necessary. Feel free to change the scripts if you would like to organize things differently.
While it is possible to organize this type of work around a set of local files, it quickly turns into unworkable mess (guess how I know). The recommendation is thus to use a database. This is not so much about the volume of the data, which in this exercise is actually not so big, but rather about the ability to crossreference and collate the information. Progesterone pipeline uses MySQL, the schema can be found in 00_progesterone_db.sql. Running
mysql -u <username> -p <passw> < 00_progesterone_db.sql
will create the database called "progesterone" for you. We will fill it as we progress though the steps below.
You may start by downloading (human and mouse) chromosome lengths and gene coordinates from UCSC using 01_chromosome_lengths_from_UCSC.py and 02_all_gene_ranges_from_UCSC.py. The scripts will stuff them in the database we have just created. On the top of 02_all_gene_ranges_from_UCSC.py you can specify the chromosome and assembly to be (down)loaded. 03_assembly_shorthands_from_UCSC.py downloads mapping between assembly and species names. This comes handy in 04_single_gene_range_from_UCSC.py where we specify gene coordinates for gene of special interest, across multiple species.
With 2 meters of DNA squished in the nucleus with 6 micrometres in diameter, we expect that the packing is quite complex. What we know in addition, is that the packing is not random. Rather, the genetic material is organized into domains, TADs, that stay localized in space, even as they keep rearranging internally, and switching their positions within the nucleus, reviewed in Merkenschlager & Nora. The implication would be that any transcription factor (TF) binding site therein is potentially regulating any (or all) genes therein. This means that we can look for the regulating TF binding sites as far as ~MBp away from the promoter, an idea that would be an anathema half a decade back. In addition it opens a happy possiblity that genes are not under a single TF site control, but under control of a number of them, in a stochastic way, with the probablity of TF having an effect being proportional to the number of TF binding sites in a TAD.
Where on the chromosome, then, is the TAD that my gene belongs to? The idea repeatedly appears in the literature that TAD boundaries are conserved across cell types and species (for example, Dixon et al review, Moll Cell 2016). That would be useful, beacuse with unversal definetion of TADs we could write very many scripts with very many purposes, having looked up the TAD definition only once.
06_tads_overview.py explores that possibility - with mixed results.
You will need to download TAD files from the Yue lab page (we suggest sticking with hg19 throughout the pipeline - use link named 'TADs in hg19'). Adjust the dirpath in 06_tads_overview.py accordingly. This script groups TAD intervals that appear repeatedly in different experiments, in the hope of finding those that are supposedly conserved across different cell types. It seems that one could make some progress looking for regions that are rarely assigned to a domain, and thus delineate the topmost division in a TAD hierarchy. The number of these divisions, however, appears to be an order of magnitude smaller than the number of TADs typically reported in each experiment.
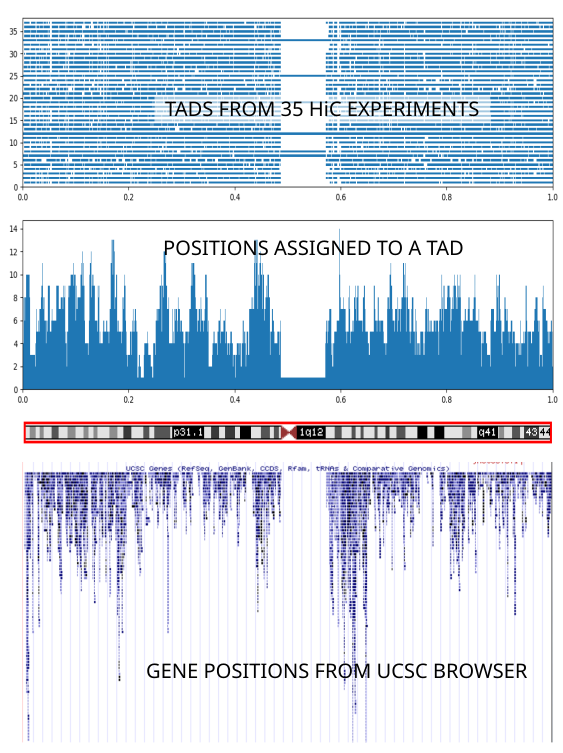
08_tads_pic.py contains basic code to illustrate the reported TAD domains. It relies on the output produced by 06_tads_overview.py. It uses Matplotlib; note that in Matplotlib you can zoom into any region of the graph you are interested in. Here are the TADS for human chromsome 1, from 35 different experiments from the Yue lab collection:
The whole chromosome length has been rescaled to the range [0,1]. If you squint a little you can see that the basic TAD structure corresponds to the regions of densest gene occupation region (middle and bottom panels) - careful with the interpretation of this graph: see 08_tads_pic.py:plot_2()). However the individual TAD assignments (top panel; each horizontal level corresponds to one experiment) vary widely between different experiments. Therefore we choose to stick with a single experiment (Homo sapiens endometrial microvascular endothelial cells) because the cell type it uses matches most closely the type of cells we are interested in.
Caveat: on some OS/graphics card setups 08_tads_pic.py may crash. It is the matter of Matplotlib rather than the script itself.
Provided you have downloaded gene ranges using 02_gene_ranges_from_UCSC.py and installed bed file containig definition of TADs you would like to stick with (this pipeline originally used this), 10_store_tads.py will store it in the db. After that 12_gene_tad.py will quickly find the coordinates of the TAD that your gene belongs to.
Note: in all cases we are actually looking at ChIPSeq regions, which are much larger than the actual TF binding site. We will narrow down each of these regions later in the pipeline.
14_tfbs_from_UCSC.py will download ChIPSeq information from ENCODE deposited in UCSC. The contents of the relevant database table are described here. This information relates to human only. Also some TFs are notably absent - progesterone for example :}. When running the script you can choose (from the command line) whether to run it for a region or for a whole chromosome.
15_tfbs_UCSC_sources.py will output the contents of the table informatively named wgEncodeRegTfbsClusteredInputsV3. This is the closest yours truly could get to finding the actual information about the ENCODE experiments behind each ChIPSeq entry. At this point, to get the actual ENCODE experiment ID you will have to get creative on the ENCODE search page and use the combined search on cell type and treatment or some such. Note that the experiment numbers output here refer to the expNum field in the tables produced by 14_tf_binding_sites_from_UCSC.py.
16_UCSC_sources_to_ENCODE.py is a patch to systematically connect that info inside a database table.
For description of bed format click here.
17_tfbs_from_local_bed.py will produce equivalent output to 14_tf_binding_sites_from_UCSC.py, but starting from a local list of bedfiles (the files should refer to ChIPSeq experiments, and can be found on GEO and ENCODE pages, for example, or you may use your own source.) 17_tfbs_from_local_bed.py will take as the input on the command line the path to the data directory and tsv table containing some basic input file meta-data.
The data directory should preferably contain bed files, grouped by experiment id, like this:
├── GSE36455
│ ├── GSE36455_series_matrix.tsv
│ ├── GSM894053_WT_vehicle.ER_peaks.bed
│ ├── GSM894054_WT_E2.ER_peaks.bed
│ └── README
└── GSE62475
├── GSE62475_series_matrix.tsv
├── GSM1527528_pgra-v-input-macs_peaks.bed
├── GSM1527529_pgrb-v-input-macs_peaks.bed
└── README
The tsv should contain tab separated columns of the form
organism | gene or chr name | TAD exp id | TF name | source | experiment id | agonist file bed | control/vehicle file bed | assembly.
Control file is optional - but the control peaks will be subtracted from the peaks in the presence of agonist,
if available. TAD experiment is ignored if a chromosome name is given.
If TAD exp id is given, TAD region encompassing the gene will be used as target range.
Otherwise 1Mbp on each side of the gene will be used.
There are some special considerations to be taken in account here:
17_tfbs_from_local_bed.py expects "bed" format, which here means that the columns are tab separated, the first column is chromosome number (possibly prefixed by 'chr'), and the following two are region start and region end. The format is still not universally accepted as the standard, so in GEO for example you can find data deposited in assorted ad hoc formats. If the file is not too far from that format, you can perhaps help yourself out with linux cut command, see also here for tab delimiter handling with cut. For example
cut -d$'\t' -f3-5 GSM857545_1_PR_oil_s_4_aligned.tsv > GSM857545_1_PR_oil_s_4_aligned.bed
Progesterone pipeline takes hg19 and mm9 as its reference assemblies. In GEO repositories there is usually a file called *_series_matrix.txt (which is actually a tsv file), where this info can be found. You may grep for hg or mm to see which one is referred to. If it is not hg19 for human or mm9 for mouse, the coordinates need to be translated. (In Windows you might try using a spreadsheet program to reformat the file. Just make sure you do not have too many in your bed file). Include that information in the table you pass to 17tfbs_from_local_bed.py and it will translate the coordinates for you, provided the following two resources: CrossMap, a copy of which is included in this distribution in the utils directory, and transformation chain files). The script will inform you if it cannot find these files in the place where it expects them.
Is the TAD that contains 'my' gene privileged in the number of TF binding sites (of some particular TF) it harbors, or are the TF binding sites perhaps distributed equally across all TADs? The latter case might imply that the TF binding sites we are finding are just distributed by some random process - this would raise an awkward possibility that we are just looking at noise.
18_tfbs_distribution.py counts the number of TF binding sites for each TAD. For Gnuplot users, an input like 19_tfbs_distribution.gplt can be used to produce a quick and dirty visualization of the TFs-per-TAD histogram. The output from 17_tfbs_distribution.py can also be used to check correlation between TAD length and the number of TF binding sites therein (mercifully, there does not seem to be any).
20_motif_finder_exercise.py illustrates basic usage of a position weight matrix (courtesy of JASPAR) together with Motif from Biopython to find a motif within a given range.
23_motif_in_chipseq_region.py will scan each chipseq range from a file produced by 14_tf_binding_sites_from_UCSC.py or 16_tfbs_from_local_bed.py and report all motifs scoring beyond certain cutoff. For such motifs it will proceed to look for the alignment with other vertebrates. (The idea with using all vertebrates is to see how far in back in the evolution the conservation can be tracked, or to eliminate motifs that are taxonomy branches that do not exhibit the same regulatory behavior; you may go for pairwise alignments with smaller number od species.) Note: extracting the alignment from maf files is a very slow process. You may turn it off in the first pass, or just choose not to use it at all, especially since the required maf files take a lot of space and long time to download.
HiC experiment we came across here have their results stored in HDF5 format, so this is the place where you might want to install h5py. We will investigate what is the 'intensity' (a number proportional to the probability that two HiC regions meet in time and space) of interaction between HiC region containing the promoter of the regulated gene, and regions containing the TF binding sites.
24_hdf5_exercise.py contains some warmup exercised in hdf5 file manipulation, and 25_hdf5_exercise2_normalization.py checks whether the rows and columns of the interaction matrix are normalized (or 'balanced'; see Lajoie, Dekker and Kaplan).
26_chromatin_interactions_from_ENCODE.py will create a mapping between TF binding sites and HiC regions, sorted by the 'interaction strength' with the HiC region that contains the promoter of the regulated gene. The hardcoded paths at the top of the main() are kinda gross, fixing it is on the TODO list.
In some cases data from ATAC-Seq experiment are available, giving some indication about the (change in) chromatin accessibility. For the case this pipeline was originally intended for, it can be found here, the information about chromatin accessibility change upon progesteron administration in human endometrial stromal cells. 30_accessible_regions.py pools it together, in the form of a Matplotlib plot, with the coordinates of our gene of interest and of TF binding sites for two TFs: ESR1 and PGR.
The final goal of the whole exercise is to collate data from all these disparate sources we started with. 35_report.py is one possibility, but this is definitely one you should rewrite or replace with something that more closely fits your needs.
Print the table you have made in the last step, and take it to a competent experimentalist. (S)he might be able to help.
- Some files still have 'human', 'mouse', 'ESR1' or 'PGR' etc hardcoded. Move to command line.
- Pull (pool?) all reference choices (reference assemblies, reference species for the alignment) in a single place.
- Get rid of CrossModule.py and use cmmodule directly.