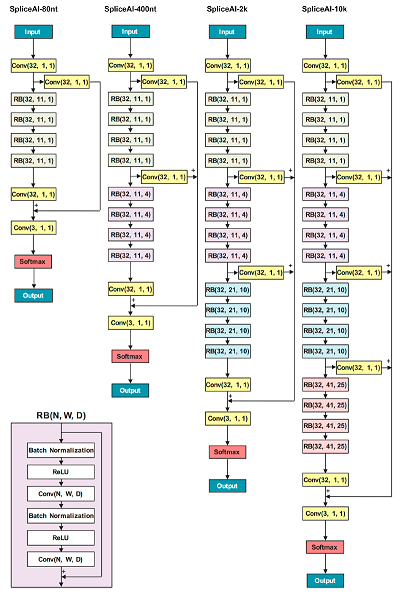
Implementation of SpliceAI, Illumina's deep neural network to predict variant effects on splicing, in PyTorch. You can find the Illumina's official implementation here.
pip install spliceai-pytorchimport torch
from spliceai_pytorch import SpliceAI
model_80nt = SpliceAI.from_preconfigured('80nt')
# model_400nt = SpliceAI.from_preconfigured('400nt')
# model_2k = SpliceAI.from_preconfigured('2k')
# model_10k = SpliceAI.from_preconfigured('10k')
# ... training ...
x = torch.randn([1, 4, 80 + 5000]) # Predicts Donor/Acceptor probs only for core 5000nt region.
probs = model_80nt(x) # (1, 5000, 3)First, download 'SpliceAI train code' directory from here and unzip it to spliceai_train_code directory.
Also, download human reference genome (version hg19) to spliceai_train_code/reference directory.
Then, run the following command to generate train/test sets after moving into spliceai_train_code/Canonical.
# Before running `grab_sequence.sh`,
# make sure that the variable CL_max is configured properly in `constants.py` (80, 400, 2000 or 10000)
chmod 755 grab_sequence.sh
./grab_sequence.sh
# Requires Python 2.7, with numpy, h5py, scikit-learn installed
python create_datafile.py train all # ~4 miniutes, creates datafile_train_all.h5 (27G)
python create_datafile.py test 0 # ~1 minute, creates datafile_test_0.h5 (2.4G)
python create_dataset.py train all # ~11 minutes, creates dataset_train_all.h5 (5.4G)
python create_dataset.py test 0 # ~1 minute, creates dataset_test_0.h5 (0.5G)$ python -m spliceai_pytorch.train --model 80nt \ # 80nt, 400nt, 2k, 10k
--train-h5 spliceai_train_code/Canonical/dataset_train_all.h5 \
--test-h5 spliceai_train_code/Canonical/dataset_test_0.h5 \
--use-wandb # Optional, for logging.Currently on the reproduction of Figure 1E. Results are as below, and you can view model training logs here (W&B).
NOTE: Target results are from ensemble of 5 models, while reproduced results are from a single model.
| Model | Top-k acc. (target) | PR-AUC (target) | Top-k acc. (reproduced) | PR-AUC (reproduced) |
|---|---|---|---|---|
| SpliceAI-80nt | 0.57 | 0.60 | 0.54355 | 0.56435 |
| SpliceAI-400nt | 0.90 | 0.95 | 0.87265 | 0.93160 |
| SpliceAI-2k | 0.93 | 0.97 | 0.9083 | 0.9541 |
| SpliceAI-10k | 0.95 | 0.98 | 0.9286 | 0.96475 |
@article{jaganathan2019predicting,
title={Predicting splicing from primary sequence with deep learning},
author={Jaganathan, Kishore and Panagiotopoulou, Sofia Kyriazopoulou and McRae, Jeremy F and Darbandi, Siavash Fazel and Knowles, David and Li, Yang I and Kosmicki, Jack A and Arbelaez, Juan and Cui, Wenwu and Schwartz, Grace B and others},
journal={Cell},
volume={176},
number={3},
pages={535--548},
year={2019},
publisher={Elsevier}
}