Phertilizer: growing a clonal tree from ultra-low coverage single-cell DNA sequencing data of tumors
For more details, see: https://doi.org/10.1101/2022.04.18.488655.
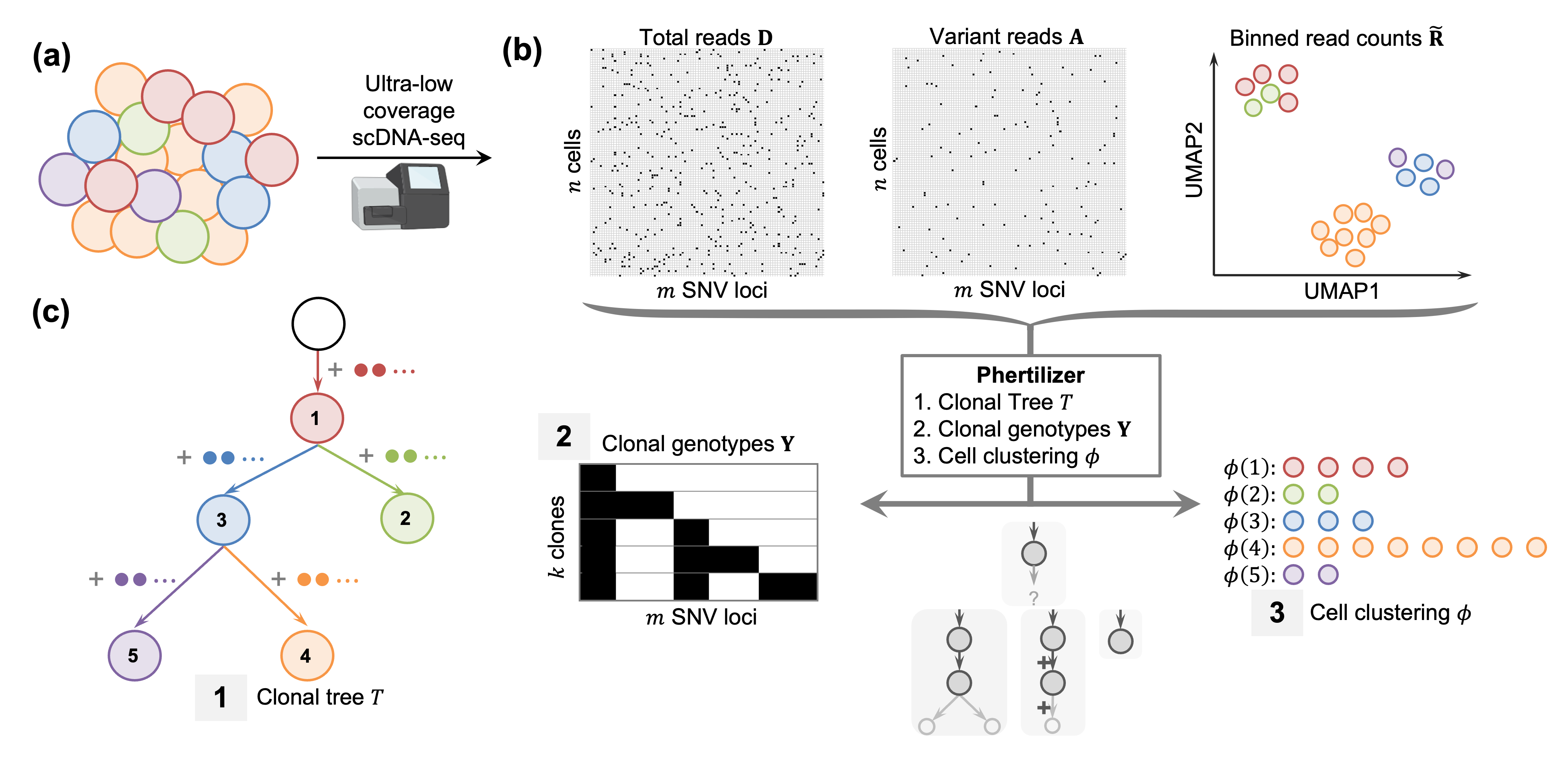 Phertilizer infers a clonal tree with SNV genotypes and a cell clustering given ultra-low coverage single-cell sequencing data.
(a) A tumor is composed of groups of cells, or clones with distinct genotypes.
(b) Ultra-low coverage single-cell DNA sequencing produces total read counts and variant read counts for n cells and m SNV loci, and low dimension embedding for the same cells for an input set of binned read counts.
(c) Phertilizer infers a clonal tree, SNV genotypes and cell clustering with maximum posterior probability.
Phertilizer infers a clonal tree with SNV genotypes and a cell clustering given ultra-low coverage single-cell sequencing data.
(a) A tumor is composed of groups of cells, or clones with distinct genotypes.
(b) Ultra-low coverage single-cell DNA sequencing produces total read counts and variant read counts for n cells and m SNV loci, and low dimension embedding for the same cells for an input set of binned read counts.
(c) Phertilizer infers a clonal tree, SNV genotypes and cell clustering with maximum posterior probability.
This is the Phertilizer code repository. The Phertilizer data repository is located at https://github.com/elkebir-group/phertilizer_data.
- Clone the repository
$ git clone https://github.com/elkebir-group/phertilizer.git
- Install phertilizer using pip
$ pip install ./
- python3 (>=3.7)
- numpy
- pandas
- numba
- scipy
- networkx(<=3.1)
- scikit-learn(>=1.1.2)
- pygrahpviz
- umap
The input for Phertilizer consists of two text based file:
- A tab or comma separated dataframe with unlabeled columns: |chr | snv | cell | alternate base | variant_reads | total_reads |
- A tab or comma separated dataframe for binned reads counts for tumor cells with labeled columns: |cell | bin1 | bin2 | ... | binb |
Note: cell ids in binned read count file should exactly match cell ids in the variant reads dataframe
See example/input for examples of all input files.
The ouput file options include:
- A png of the clonal tree with maximum posterior probability
- A text file containing the edge list of the tree
- A dataframe mapping cells to nodes
- A dataframe mappping SNVs to nodes
- A pickle file of the clonal tree with maximum posterior probability
- A pickle file containing a ClonalTreeList of all enumerated clonal trees
See example/output for examples of output files 1 through 4.
$ phertilizer -h
usage: phertilizer [-h] -f FILE --bin_count_data BIN_COUNT_DATA [-a ALPHA] [-j ITERATIONS] [-s STARTS] [-d SEED] [--radius RADIUS] [-c COPIES]
[--runs RUNS] [-g GAMMA] [--min_obs MIN_OBS] [-m PRED_MUT] [-n PRED_CELL] [--post_process] [--tree TREE]
[--tree_pickle TREE_PICKLE] [--tree_path TREE_PATH] [--tree_list TREE_LIST] [--tree_text TREE_TEXT] [--likelihood LIKELIHOOD]
[--embedding EMBEDDING] [--no-umap] [--low_cmb LOW_CMB] [--high_cmb HIGH_CMB] [--nobs_per_cluster NOBS_PER_CLUSTER]
optional arguments:
-h, --help show this help message and exit
-f FILE, --file FILE input file for variant and total read counts with unlabled columns: [chr snv cell base var total]
--bin_count_data BIN_COUNT_DATA
input binned read counts with headers containing bin ids or embedding dimensions
-a ALPHA, --alpha ALPHA
per base read error rate
-j ITERATIONS, --iterations ITERATIONS
maximum number of iterations
-s STARTS, --starts STARTS
number of restarts
-d SEED, --seed SEED seed
--radius RADIUS
-c COPIES, --copies COPIES
max number of copies
--runs RUNS number of Phertilizer runs
-g GAMMA, --gamma GAMMA
confidence level for power calculation to determine if there are sufficient observations for inference
--min_obs MIN_OBS lower bound on the minimum number of observations for a partition
-m PRED_MUT, --pred-mut PRED_MUT
output file for mutation clusters
-n PRED_CELL, --pred_cell PRED_CELL
output file cell clusters
--post_process indicator if post processing should be performed on inferred tree
--tree TREE output file for png (dot) of Phertilizer tree
--tree_pickle TREE_PICKLE
output pickle of Phertilizer tree
--tree_path TREE_PATH
path to directory where pngs of all candidate trees are saved
--tree_list TREE_LIST
pickle file to save a ClonalTreeList of all generated trees
--tree_text TREE_TEXT
text file save edge list of best clonal tree
--likelihood LIKELIHOOD
output file where the likelihood of the best tree should be written
--embedding EMBEDDING
filename where the UMAP coordinates should be saved after embedding binned read counts
--no-umap flag to indicate that input reads per bin file should NOT undergo additional dimensionality reduction
--low_cmb LOW_CMB regularization parameter to assess the quality of a split where CMB should <= low_cmb for parts of an extension
--high_cmb HIGH_CMB regularization parameter to assess the quality of a split where CMB should >= high_cmb for parts of an extension
--nobs_per_cluster NOBS_PER_CLUSTER
regularization parameter on the median number of reads per cell/SNV to accept extension
Here we show an example of how to run Phertilizer.
The input files are located in the example/input directory.
$ phertilizer -f example/input/variant_counts.tsv \
--bin_count_data example/input/binned_read_counts.csv \
--tree example/output/tree.png \
--tree_text example/output/tree.txt \
-n example/output/cell_clusters.csv \
-m example/output/SNV_clusters.csv \
-s 3 -j 10 --post_process
This command generates output files tree.png,tree.txt, cell_clusters.csv, and SNV_clusters.csv in directory example/output.