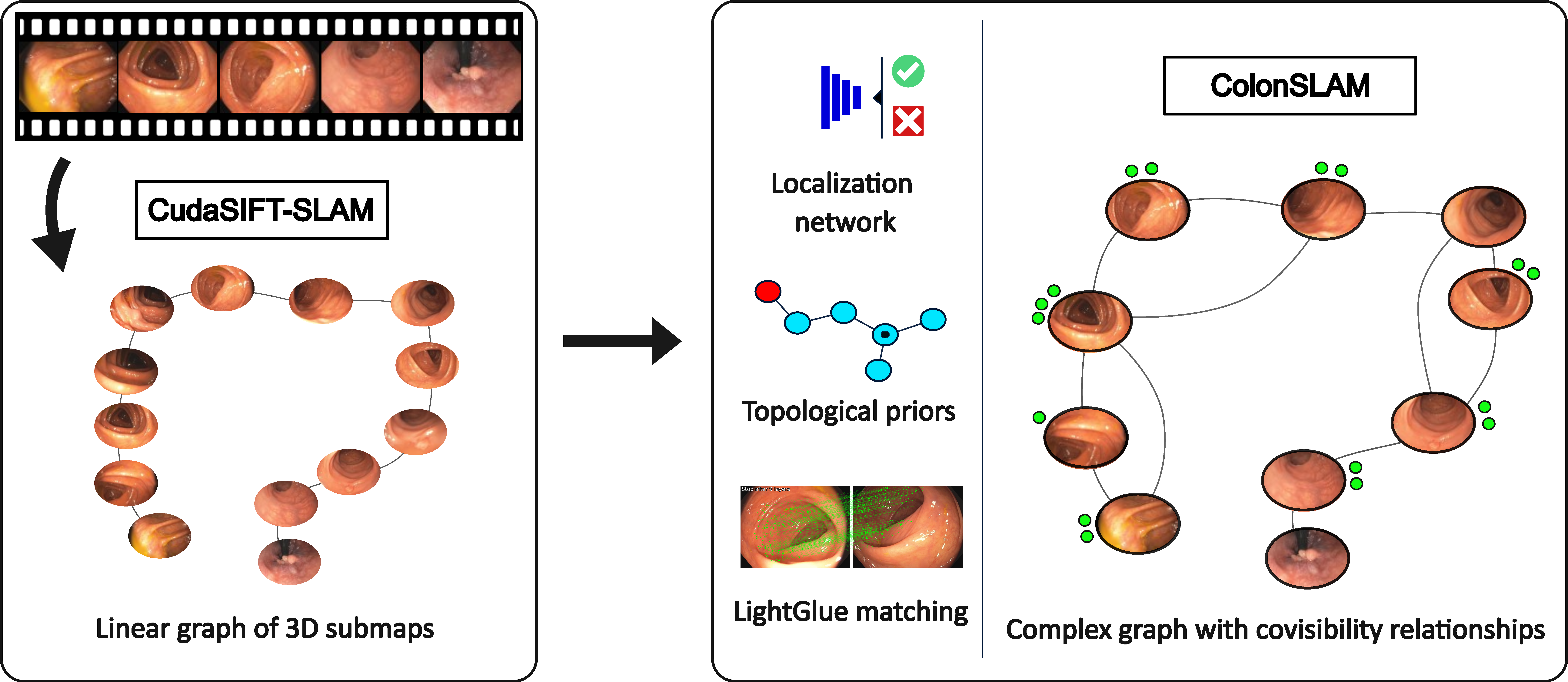
ColonSLAM is a metric-topological SLAM algorithm able to build a topological graph starting from the metric submaps built by CudaSIFT-SLAM. Additionally, it can localize a second sequence of the same patient against the previously built topological map.
To install the ColonSLAM environment, simply use conda as:
conda create -f environment.ymlFirst, download the trained models, which can be found here.
The images used for evaluation can be found here.
To run the topological SLAM run the following command:
cd ColonSLAM
python slam.py --resume=[PATH_TO_MODELS]/endofm_cls_0_0_224_schedule_resize_none/best_model.pth --use_lightglue --experiment_name=colonslam --sim_threshold=0.95 --use_mlp --matches_threshold=100 --window_size=5 --voting_threshold=0.20 --filterTo run the map reuse experiment, run the following command:
cd ColonSLAM
python slam_reuse.py --resume=[PATH_TO_MODELS]/endofm_cls_0_0_224_schedule_resize_none/best_model.pth --experiment_name=colonslam-reuse --sim_threshold=0.80 --use_mlp --mode=slam --voting_threshold=0.15 --sequence_map 027 --start_processing 34 --sequence_loc 035 --window_size=5 --filter Javier Morlana, Juan D. Tardós J.M.M. Montiel, Topological SLAM in colonoscopies leveraging deep features and topological priors, MICCAI 2024. PDF
@inproceedings{morlana2024topological,
title={Topological SLAM in colonoscopies leveraging deep features and topological priors},
author={Morlana, Javier and Tard{\'o}s, Juan D and Montiel, Jos{\'e} MM},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={733--743},
year={2024},
organization={Springer}
}