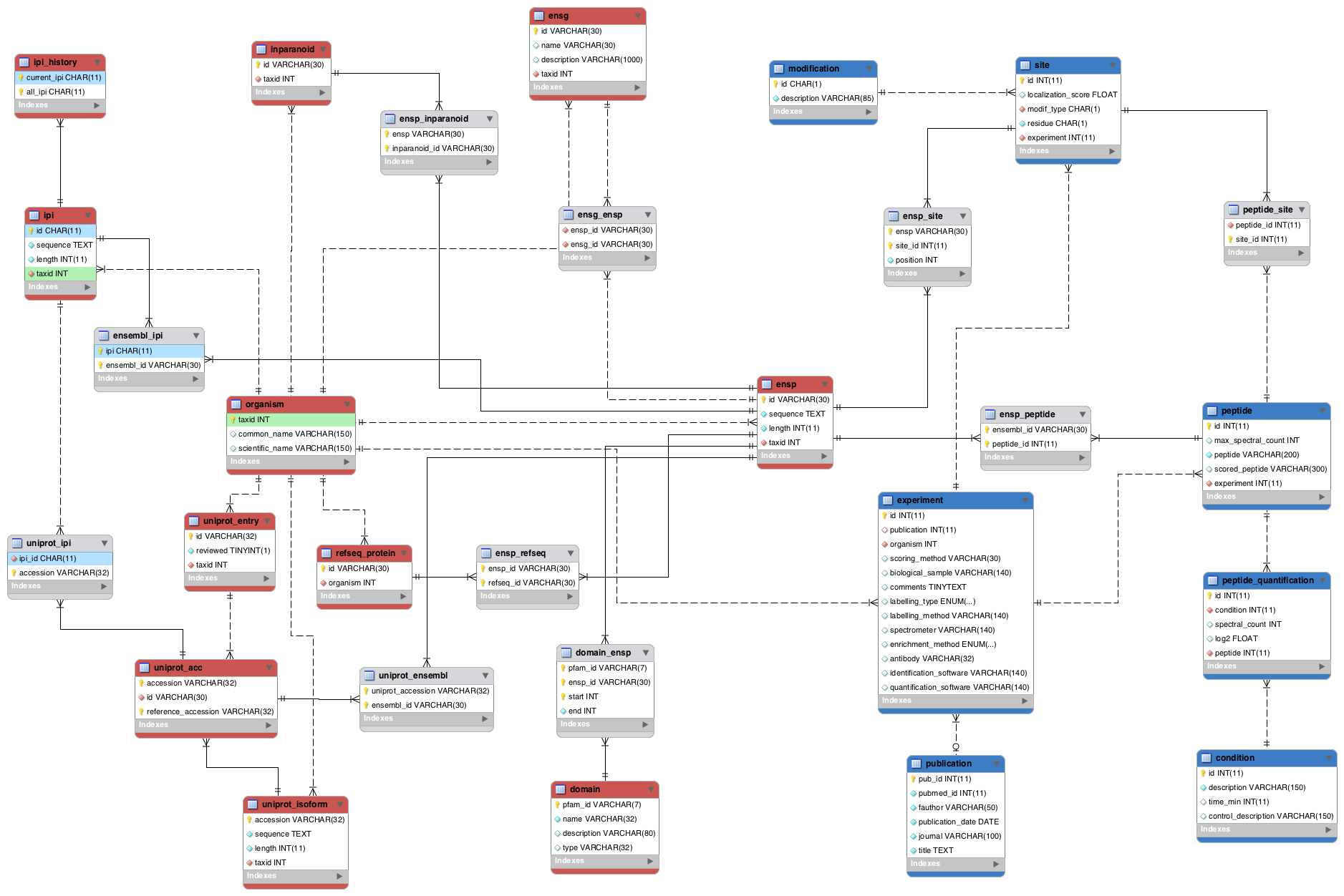
This project contains all the necessary code to create and fill a database of post-translational modifications. The database will contain the quantitative and non-quatitative ptms available as well as the necessary information to map it through different IDs.
- Requirements:
Check out the Makefile to be sure of the external databases
necessary to fill the database.
-
Dependencies:
-
perl - packages: DBI, DBD::MySQL, LWP
-
R - packages: RMySQL, (devtools?)
-
wget - Not installed in OSX
-
GNU sed (OSX default sed doesn't work)
-
This repository can act also as a submodule of other projects intended to use the ptmdb.
First, you must define the organisms that will be present in the
database in the organism.csv file. This file consists of eight
tab-delimited fields:
- the common name (i.e. "human")
- the scientific name ("Homo sapiens")
- the taxonomic ID ("9606")
- the IPI ID ("ipi.HUMAN" or "NA" if it doesn't exist)
- the Ensembl FTP master directory in which the species directories reside ("ftp://ftp.ensembl.org/pub", "ftp://ftp.ensemblgenomes.org/pub/plants", etc.)
- the Ensembl proteome version ("GRCh37")
- the Ensembl release number ("74")
- the BioMart Database ("ENSEMBL GENES 74")
Next, you must define the classes of post-translational modifications
that will be stored in the database in the modifications.csv file.
This is a two-column, tab-delimited file consisting of the
modification abbreviation ("P") and its full name ("Phosphorylation").
You must also create the database itself and to grant privileges to a
user. Be sure to set the user and database information inside the
Makefile.
Setting up the database tables and populating them with the proteomics
data are handled via the Makefile. If your organism.csv and
modifications.csv files are correct, simply running make will
automate the whole process. This process can be broken down into
individual steps as follows:
make tables modifications xml-queries parse-histories proteomes insert-species
Thus, to only download the proteomic data, run make proteomes.
To silence some of Make's output, run it with the -s option. To
parallelise some of the processes, use the -j option (i.e. -j4
runs four parallel processes). You probably only want to parallelise
the downloads of proteomic data (make -j4 proteomes; make all).
The reference databases and all the mappings can be updated with the
next commands. The NEWDB variable contains the new database name
where all the contents will be dumped. During the process, the IDs
will change using the Ensembl ID conversion when possible. If not, uniprot
is used to translate old to new Ensembl IDs. All peptides are remapped,
and the sites are relocated on their new positions.
make updateDB NEWDB=ptmdb3
There are three clean targets in the Makefile. The first two
delete incidental data that is not necessary after the database has
been set up: clean-proteomes and clean-xml. The former deletes
all of the downloaded proteomic data while the latter deletes XML
Biomart queries. Running make clean is equivalent to make clean-proteomes clean-xml. Finally, the clean-tables target clears
data out of the database. Needless to say, none of these actions can
be undone, so use caution.
ptmdbR has been created as a subproject of ptmdb and serves as an interface between the database and R. Some of the most frequent actions such as connecting with the database or querying its content are implemented as simple functions. This framework should be useful when asking higher order questions.
Integrating ptmdbR on your on project is extremely easy. You just have to invoke the library from the proper path:
library(ptmdbR, lib.loc="<path to the libraries parent directory>")
To add or modify ptmdbR functions, you can find the code for the whole
package under src/ptmdbR/src. Note that the package code is
commented using
roxygen2. roxygen2 allows to
produce the documentation for the different functions. Be kind a keep
using it.
Once the code has been modified it's necessary to generate the updated
documentation and build the package. In order to simplify this step,
you can just run make ptmdbR in the main directory and the
compiled ptmdbR package will be created in the location specified
by PTMDBRLIBLOC in the Makefile.