HURON is a command line java application for ontology metrics calculation. Further details about the metrics are available at http://semantics.inf.um.es/huron/docs_metrics.html.
java -jar huron.jar --input <arg> [--output-long <arg>] [--output-wide <arg>] [--output-rdf <arg>] [--detailed-files <arg>] [--imports] [--threads <arg>]
Where
- --input Input file, which be an owl ontology, or a folder containing owl ontologies.
- --output-long Output tsv file with the metrics in long format with the columns 'ontology', 'metric' and 'value'.
- --output-wide Output tsv file with the metrics in wide format, where the metrics are in different columns.
- --output-detailed-rdf Output RDF file according to oquo ontology, including information about all entities in the ontology.
- --output-summary-rdf Output RDF file according to oquo ontology, including only information about the ontology.
- --detailed-files Generate a report for each metric in the folder passed as argument.
- --threads Number of threads to use in parallel.
- --imports Consider imported entities from external ontologies (import clause) when calculating the metrics.
If you want to test, modify or compile the application from the source code, you will need access to the ontoenrich-core library, used by the application to perform the analysis of lexical regularities. The source code of this library is not available yet, but it is supported by several publications:
- https://link.springer.com/chapter/10.1007/978-3-319-17966-7_25
- https://hal.archives-ouvertes.fr/hal-03155057/
You can download this library from here.
An online version of HURON is available at http://semantics.inf.um.es/huron.
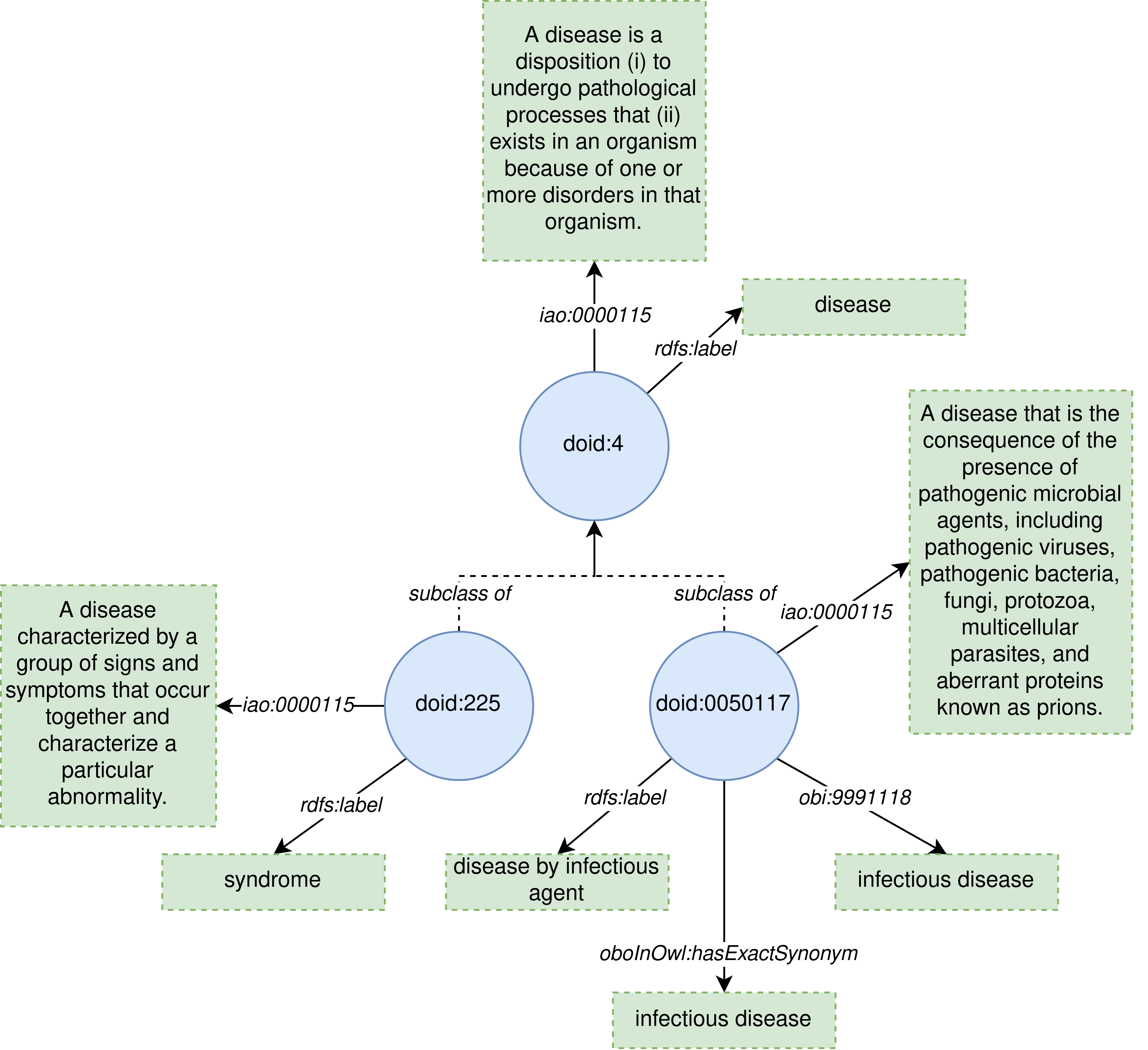
As an example of how the metrics work, lets assume the following ontology:
This ontology is an excerpt of the Human Disease Ontology consisting of 3 classes that have been annotated with some human-readable content, such as:
- Names, by using rdfs:label.
- Descriptions, by using IAO:0000115.
- Synonyms, by using oboInOwl:hasExactSynonym and obi:9991118.
This ontology is available here, and it has been used for testing the application; the corresponding JUnit test case is available here.
The ontology contains 3 classes, being each one annotated with an rdfs:label, which is considered as a name. Thus the value for the metric names per class is calculated as 3 classes over 3 names in the ontology, which results in 1 name per class.
The ontology contains 3 classes, and each one is annotated with one description through the property IAO:0000115. The value for the descriptions per class metric is calculated as 3 classes over 3 descriptions, resulting in 1 description per class.
The ontology contains 3 classes, doid:4 and doid:225 do not include any synonym, whereas doid:0050117 include two, one annotated through the property oboInOwl:hasExactSynonym, and another one through obi:9991118. Then, the ontology has a total of 2 synonyms over 3 classes, resulting in 2/3 synonyms .er class.
The ontology contains the lexical regularity `disease' because that text is exhibited in the name of the classes doid:4 (disease), and doid:0050117 (disease by infectious disease). As this lexical regularity matches exactly with the name of doid:4, the class doid:4 is a lexical regularity class. The Lexically Suggest Logically Define (LSLD) and the Systematic Naming metrics are computed over the lexical regularity classes.
The only lexical regularity class of the ontology is doid:4 (disease). This class has a total of 2 subclasses, being one of them doid:0050117 (disease by infectious disease), which exhibit the lexical regularity "disease", and the other one doid:225 (syndrome), which does not exhibit the lexical regularity "disease" in its name. Thus, the ontology has a systematic naming value of 1 class exhibiting the lexical regularity of the parent lexical regularity class over a total of 2 classes that are subclass of lexical regularity classes, resulting in 0.5.
The name of the lexical regularity class doid:4 (disease) is exhibited only by the class doid:0050117 (disease by infectious disease), which is also semantically related with doid:4 (disease) through a subclass axiom. Then, the LSLD value is the ratio between the number of classes that exhibit the lexical regularity of a lexical regulrity class, and the classes that are semantically related with that lexical regularity class. In this case, we only have doid:0050117 (disease by infectious disease) exhibiting the lexical regularity "disease" of the lexical regularity class doid:4 (disease), which is also semantically related with that class, thus resulting a value of 1 for the LSLD metric.