NIH Accelerating Medicines Partnership (AMP) Phase 1
This repo provides the Data availability, Source code, Website for our work on using single-cell transcriptomics and proteomics data to define inflammatory cell states in autoimmune disease - rheumatoid arthritis.
The published paper can be viewed and cited:
Zhang, F., Wei, K., Slowikowski, K., Fonseka, C.Y., Rao, D.A., et al. Defining Inflammatory Cell States in Rheumatoid Arthritis Joint Synovial Tissues by Integrating Single-cell Transcriptomics and Mass Cytometry. Nature Immunology, 2019.
The raw data of this study are available at:
| Database | Link with accession code | Data type |
|---|---|---|
| ImmPort | SDY998 | single-cell RNA-seq, mass cytometry, bulk RNA-seq, flow cytometry, clinical and histology |
| dbGAP | phs001457.v1.p1 | single-cell RNA-seq and mass cytometry |
Send us (fanzhang@broadinstitute.org or jmears@broadinstitute.org) an email if you have any quesitons or requests for data download.
cd ~/work/
git clone git@github.com:immunogenomics/amp_phase1_ra.git
cd amp_phase1_raThe files in the repo are organized as follows:
.
├── R
|── data
data/ has Excel sheets with sample metadata and RData files with processed data ready for analysis.
R/ has code for analysis and creating figures:
-
Classify tissue samples using Mahalanobis distance:
R/optimal_lymphocyte_threshold.R -
Integrate bulk with single-cell RNA-seq:
R/scRNAseq_bulkRNAseq_integrative_pipeline.R -
Cluster and disease association test using mass cytometry:
R/Tcell.SNE.densVM.server.R,R/Tcell.MASC.R -
Identify cluster marker genes:
R/cluster_marker_table.R,R/limma_differential_bulk.R -
Functions for PCA, densisty analysis, etc:
R/pure_functioins.R -
Visualize results:
R/cytof_results_plot.R,plot_cluster_markers.R, etc -
More
Send us (fanzhang@broadinstitute.org) an email if you have any quesitons for the analysis.
Feel free to check out the websites and search your favorite genes:
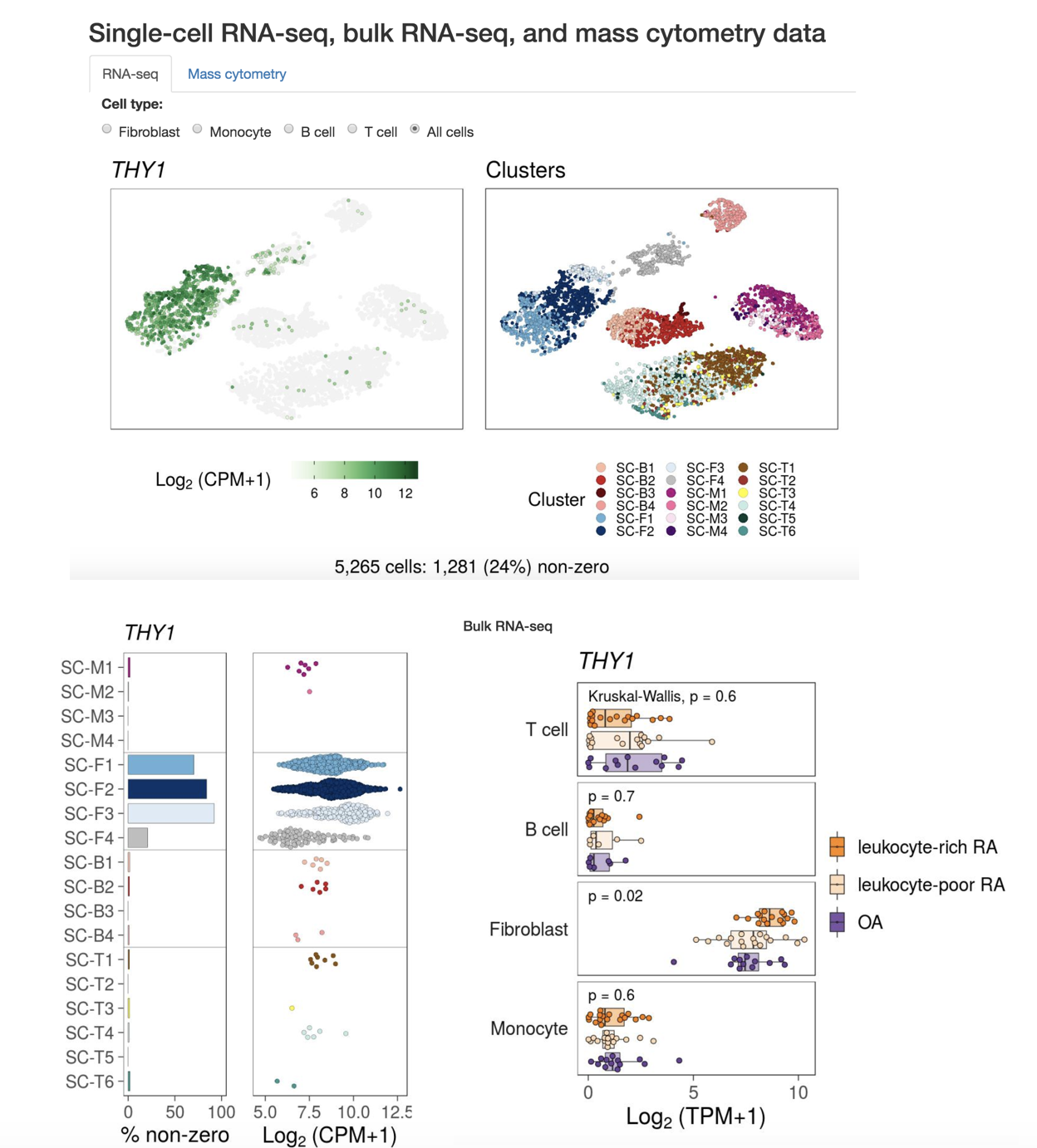
- Shiny app: view single-cell RNA-seq, bulk RNA-seq, and mass cytometry data for rheumatoid arthritis data.
- UCSC Cell Browser: view single-cell RNA-seq datasets: 1 rheumatoid arthritis dataset and 2 lupus datasets.
- Broad Institue Single Cell Portal: view single-cell RNA-seq datasets: 1 rheumatoid arthritis datset and 2 lupus datasets.
For example, get everyting in one page using Shiny app:
Send us (fanzhang@broadinstitute.org, kslowikowski@gmail.com, or jmears@broadinstitute.org) an email if you have any quesitons.