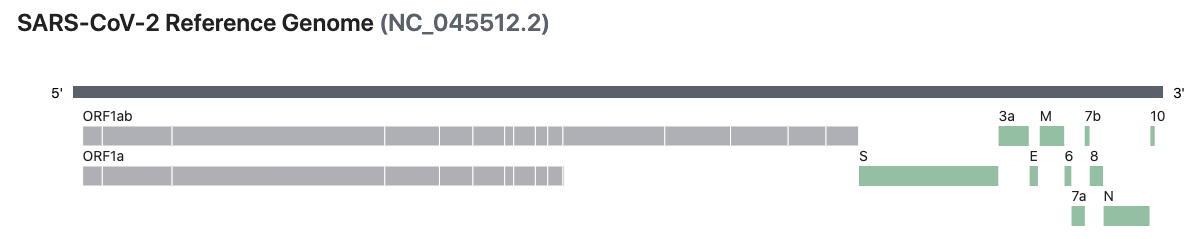
The Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an RNA virus currently causing the 2019–20 coronavirus pandemic. This repository contains my analysis code and notes for my analysis of SARS-CoV-2. My hope is that some of this work will be useful for researchers currently working on the analysis of SARS-CoV-2.
For more information see my related blog posts:
- https://davetang.org/muse/2020/03/05/sequence-analysis-sars-cov-2/
- https://davetang.org/muse/2020/03/06/sequence-analysis-of-sars-cov-2-part-2/
- https://davetang.org/muse/2020/03/12/sequence-analysis-of-sars-cov-2-part-3/
I rely on Conda (a lot) to install the tools needed to perform my analyses. I have written a short introduction to Conda that may be useful if you have never used it before. Please install Miniconda if you haven't already. Once you have Conda, run the command below to install all the necessary tools.
conda env create --file environment.yml
conda activate sars_cov_2The NCBI Datasets project has developed a command-line tool, datasets, that is used to query and download biological sequence data across all domains of life from NCBI databases.
wget https://ftp.ncbi.nlm.nih.gov/pub/datasets/command-line/LATEST/linux-amd64/datasets -O bin/datasets
chmod 755 bin/datasetsDownload Coronavirus genomes using datasets.
mkdir raw/genome
today=$(date +%Y%m%d)
bin/datasets download virus genome tax-name sars2 --filename raw/genome/sars2.${today}.zip7,031 genome sequences as of Mon Jul 20 14:32:46 JST 2020.
cat genomic.fna | grep "^>" | wc -l
7031Look for MN908947.
cat genomic.fna | grep MN908947
>MN908947.3 Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome
>MT576029.1 Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/human/ESP/2019-nCoV-MN908947-cOVID-96_19/2020, complete genomeThe download virus protein command downloads complete protein sequences (excluding partial sequences) and annotation data as a zip file in the BDBag (Big Data Bag) format.
bin/datasets download virus protein S --filename raw/SARS2-spike.zipThe default protein dataset for a given protein includes the following for all complete SARS2 RefSeq and GenBank genomes:
- amino acid sequences in FASTA (.faa) format
- protein structures in PDB (.pdb) format
- nucleotide coding (CDS) sequences in FASTA (.fna) format
- data report containing taxonomy, isolate, host and other metadata (
data_report.yaml) - annotation and amino acid sequences in the GenPept flat file format (
protein.gpff) - a README containing details on sequence file data content and other general information (
virus_dataset.md) - a list of files and file types (
dataset_catalog.json)
|-- ncbi_dataset
| |-- bag-info.txt
| |-- bagit.txt
| |-- data
| | |-- cds.fna
| | |-- data_report.yaml
| | |-- dataset_catalog.json
| | |-- pdb
| | | |-- 6VYB.pdb
| | | |-- 6VYO.pdb
| | | |-- 6W37.pdb
| | | |-- 6W4H.pdb
| | | |-- 6W9C.pdb
| | | |-- 6W9Q.pdb
| | | |-- 6WEY.pdb
| | | |-- 6WJI.pdb
| | | |-- 6WLC.pdb
| | | |-- 7BQY.pdb
| | | `-- 7BV2.pdb
| | |-- protein.faa
| | |-- protein.gpff
| | `-- virus_dataset.md
| |-- fetch.txt
| |-- manifest-md5.txt
| `-- tagmanifest-md5.txt
`-- README.mdExtract only the amino acid FASTA sequences.
for file in $(ls raw/protein/SARS2*.zip); do
base=$(basename $file .zip);
unzip -jp $file ncbi_dataset/data/protein.faa | gzip > raw/protein/${base}.faa.gz
done
zcat raw/protein/SARS2.20200720.S.faa.gz | grep "^>" | wc -l
7015Number of unique sequences.
for file in $(ls raw/protein/*.faa.gz); do
echo $file;
script/unique_seq.pl -f $file | wc -l;
done
raw/protein/SARS2.20200720.E.faa.gz
30
raw/protein/SARS2.20200720.M.faa.gz
95
raw/protein/SARS2.20200720.N.faa.gz
360
raw/protein/SARS2.20200720.nsp10.faa.gz
36
raw/protein/SARS2.20200720.nsp11.faa.gz
7
raw/protein/SARS2.20200720.nsp13.faa.gz
254
raw/protein/SARS2.20200720.nsp14.faa.gz
307
raw/protein/SARS2.20200720.nsp15.faa.gz
166
raw/protein/SARS2.20200720.nsp16.faa.gz
146
raw/protein/SARS2.20200720.nsp1.faa.gz
134
raw/protein/SARS2.20200720.nsp2.faa.gz
388
raw/protein/SARS2.20200720.nsp3.faa.gz
862
raw/protein/SARS2.20200720.nsp4.faa.gz
198
raw/protein/SARS2.20200720.nsp5.faa.gz
118
raw/protein/SARS2.20200720.nsp6.faa.gz
120
raw/protein/SARS2.20200720.nsp7.faa.gz
31
raw/protein/SARS2.20200720.nsp8.faa.gz
69
raw/protein/SARS2.20200720.nsp9.faa.gz
48
raw/protein/SARS2.20200720.ORF10.faa.gz
28
raw/protein/SARS2.20200720.orf1ab.faa.gz
5773
raw/protein/SARS2.20200720.orf1a.faa.gz
3756
raw/protein/SARS2.20200720.ORF3a.faa.gz
290
raw/protein/SARS2.20200720.ORF6.faa.gz
58
raw/protein/SARS2.20200720.ORF7a.faa.gz
88
raw/protein/SARS2.20200720.ORF7b.faa.gz
1
raw/protein/SARS2.20200720.ORF8.faa.gz
99
raw/protein/SARS2.20200720.RdRp.faa.gz
342
raw/protein/SARS2.20200720.S.faa.gz
779The page https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ contains a list of SARS-CoV-2 sequences. Download latest list of SARS-CoV-2 nucleotide sequence IDs.
outfile=acc.$(date +%F).txt.gz
wget https://www.ncbi.nlm.nih.gov/sars-cov-2/download-nuccore-ids/ -O - | gzip > raw/$outfile
zcat raw/$outfile | wc -l
10001We can use efetch to download an assembled SARS-CoV-2 genome sequence in various formats.
mkdir raw
efetch -db sequences -format genbank -id MN908947 > raw/MN908947.genbank
efetch -db sequences -format fasta -id MN908947 > raw/MN908947.fa
efetch -db sequences -format fasta_cds_aa -id MN908947We can use esearch to query the number of sequences associated with the term "coronavirus". There were 41,874 results as of 2020/03/01.
esearch -db nuccore -query coronavirus
<ENTREZ_DIRECT>
<Db>nuccore</Db>
<WebEnv>NCID_1_121026101_130.14.22.33_9001_1583054896_904858778_0MetA0_S_MegaStore</WebEnv>
<QueryKey>1</QueryKey>
<Count>41874</Count>
<Step>1</Step>
</ENTREZ_DIRECT>We can pipe the output from esearch to efetch to fetch all sequences.
today=$(date "+%Y%m%d")
time esearch -db nuccore -query coronavirus | efetch -db sequences -format fasta > raw/coronavirus_$today.fa
real 10m39.511s
user 0m53.739s
sys 0m7.705s
cat raw/coronavirus_$today.fa | grep "^>" | wc -l
43581
cat raw/coronavirus_$today.fa | grep MN908947
>MN908947.3 Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genomeI wrote a simple Perl script to calculate the length of the FASTA sequences.
today=$(date "+%Y%m%d")
script/fasta_stats.pl -f raw/coronavirus_$today.fa | gzip > result/coronavirus_${today}_stat.txt.gzCreate BLAST database using the sequences associated with the term "coronavirus".
mkdir db
makeblastdb -dbtype nucl \
-in raw/coronavirus_20200426.fa \
-input_type fasta \
-title coronavirus_20200426 \
-out db/coronavirus_20200426
Building a new DB, current time: 04/26/2020 09:29:19
New DB name: /Users/dtang/github/sars_cov_2/db/coronavirus_20200426
New DB title: coronavirus_20200426
Sequence type: Nucleotide
Keep MBits: T
Maximum file size: 1000000000B
Adding sequences from FASTA; added 43581 sequences in 13.0157 seconds.After creating the database we can blast the assembled genome to all the sequences we fetched to see if it matches other coronaviruses.
# -evalue <Real> - Expectation value (E) threshold for saving hits
# Default = `10'
blastn -outfmt 7 -query raw/MN908947.fa -db db/coronavirus_20200426 > result/MN908947_blast.txt
blastn -evalue 1 -outfmt 7 -query raw/MN908947.fa -db db/coronavirus_20200426 | wc -l
506
# accessions with BLAST hits
cat result/MN908947_blast.txt | grep -v "^#" | cut -f2 | sort -u | wc -l
500
cat result/MN908947_blast.txt | grep -v "^#" | cut -f2 | sort -u > result/MN908947_matched.txtThe script extract_fasta.pl will extract specific FASTA entries. Below we fetch all the sequences that our query sequence matched (500 in total).
script/extract_fasta.pl -i result/MN908947_matched.txt -f raw/coronavirus_20200426.fa > result/MN908947_matched.fa
cat result/MN908947_matched.fa | grep "^>" | wc -l
500We will calculate some simple FASTA stats on the matched sequences.
script/fasta_stats.pl -f result/MN908947_matched.fa > result/MN908947_matched_stats.txtThe script parse_outfmt7.pl simply parses the BLAST result and outputs the results in a more readable format.
script/parse_outfmt7.pl -i result/MN908947_blast.txt -p 80 -l 10000 -f raw/coronavirus_20200426.fa | lessCreate a multiple sequence alignment of MN908947 to other bat coronaviruses.
script/extract_fasta.pl -i raw/wanted.txt -f raw/coronavirus_20200426.fa > raw/wanted.fa
clustalw -infile=raw/wanted.fa
script/extract_fasta.pl -i raw/MN908947_MN996532.txt -f raw/coronavirus_20200301.fa > raw/MN908947_MN996532.fa
clustalw -infile=raw/MN908947_MN996532.faThe page https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ contains a YAML file with accession information on the SARS-CoV-2 sequences currently deposited on NCBI databases.
wget -N https://www.ncbi.nlm.nih.gov/core/assets/genbank/files/ncov-sequences.yaml -O raw/ncov-sequences.yaml
head raw/ncov-sequences.yaml
updated: 'Thursay Apr 23 15:15 2020 EST'
genbank-sequences: [
{
"accession": "NC_045512",
"accession_list": "<a href=\"https://www.ncbi.nlm.nih.gov/nuccore/NC_045512\">NC_045512</a>",
"collection_date": "2019-12",
"country": "China"
},
{
cat raw/ncov-sequences.yaml | grep "sra-run\"" | wc -l
288Did we get all the GenBank sequences using esearch and efetch? We should directly use the accessions in the YAML file as 417 sequences are missing.
cat raw/ncov-sequences.yaml | grep 'accession"' | perl -nle 's/.*"(\w+)",/$1/; print' > raw/genbank_list.txt
cat raw/genbank_list.txt | wc -l
1622
script/extract_fasta.pl -i raw/genbank_list.txt -f raw/coronavirus_20200426.fa > raw/genbank_list.fa 2> raw/genbank_list_missing.txt
cat raw/genbank_list.fa | grep "^>" | wc -l
1205
cat raw/genbank_list_missing.txt | wc -l
417Download using efetch and check out the distribution of sequence lengths.
my_accession=$(cat raw/genbank_list.txt | tr '\n' ',' | sed 's/,$//')
efetch -db sequences -format fasta -id $my_accession > raw/genbank_efetch.fa
cat raw/genbank_efetch.fa | grep "^>" | wc -l
1621
script/fasta_stats.pl -f raw/genbank_efetch.fa | gzip > result/genbank_stat.txt.gz
# get stats from https://github.com/arq5x/filo/
gunzip -c result/genbank_stat.txt.gz | cut -f2 | grep -v "Length" | stats
Total lines: 1621
Sum of lines: 45891887
Ari. Mean: 28310.8494756323
Geo. Mean: 23869.0792681778
Median: 29844
Mode: 29882 (N=208)
Anti-Mode: 64 (N=1)
Minimum: 64
Maximum: 29945
Variance: 41665645.6466825
StdDev: 6454.89315532663
# some short sequences
gunzip -c result/genbank_stat.txt.gz | sort -k2n | head -6
Accession Length A C G T Unknown
MT293547.1 64 13 16 12 23 0
MT273658.1 84 20 15 22 27 0
MT163712.1 87 20 17 23 27 0
MN938387.1 107 38 14 22 33 0
MN938388.1 107 38 14 22 33 0
cat raw/genbank_efetch.fa | grep MT293547.1
>MT293547.1 Severe acute respiratory syndrome coronavirus 2 isolate SARS-CoV-2/human/IRQ/KRD/2020 envelope protein (E) gene, partial cdsUse wget to obtain metadata of all SARS-CoV-2 raw sequences upload to the SRA. The YAML file no longer uses project accessions (SRP242226), so we will get information on each run instead (SRR10948550).
mkdir sra
# "sra-run": "SRR10948550"
wget -O sra/SRR10948550_info.csv 'http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?save=efetch&db=sra&rettype=runinfo&term=SRR10948550'Checkout metadata using csvkit.
csvcut -n sra/SRR10948550_info.csv
1: Run
2: ReleaseDate
3: LoadDate
4: spots
5: bases
6: spots_with_mates
7: avgLength
8: size_MB
9: AssemblyName
10: download_path
11: Experiment
12: LibraryName
13: LibraryStrategy
14: LibrarySelection
15: LibrarySource
16: LibraryLayout
17: InsertSize
18: InsertDev
19: Platform
20: Model
21: SRAStudy
22: BioProject
23: Study_Pubmed_id
24: ProjectID
25: Sample
26: BioSample
27: SampleType
28: TaxID
29: ScientificName
30: SampleName
31: g1k_pop_code
32: source
33: g1k_analysis_group
34: Subject_ID
35: Sex
36: Disease
37: Tumor
38: Affection_Status
39: Analyte_Type
40: Histological_Type
41: Body_Site
42: CenterName
43: Submission
44: dbgap_study_accession
45: Consent
46: RunHash
47: ReadHash
csvcut -c Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName sra/SRR10948550_info.csv | csvlook
| Run | Sample | BioSample | spots | LibraryStrategy | LibrarySource | LibraryLayout | Platform | Model | SampleType | CenterName |
| ----------- | ---------- | ------------ | ------- | --------------- | ------------- | ------------- | --------------- | ------ | ---------- | --------------------- |
| SRR10948550 | SRS6014638 | SAMN13871323 | 425,717 | RNA-Seq | GENOMIC | SINGLE | OXFORD_NANOPORE | MinION | simple | HKU-SHENZHEN HOSPITAL |
| | | | | | | | | | | |Download all metadata using SRR accessions from raw/ncov-sequences.yaml.
my_accession=$(cat raw/ncov-sequences.yaml | grep 'sra-run"' | perl -nle 's/.*"(\w+)",/$1/; print')
for acc in $my_accession; do
wget -O sra/${acc}_info.csv "http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?save=efetch&db=sra&rettype=runinfo&term=$acc"
done
# concenate into one metadata file
rm -f sra/metadata.txt
touch sra/metadata.txt
for file in `ls sra/SRR*info.csv`; do
echo $file;
csvcut -c Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName $file >> sra/metadata.txt
done
head sra/metadata.txt
Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName
SRR10902284,SRS6014638,SAMN13871323,261890,RNA-Seq,METAGENOMIC,SINGLE,OXFORD_NANOPORE,MinION,simple,UNIVERSITY OF HONG KONG
,,,,,,,,,,
Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName
SRR10903401,SRS6007144,SAMN13872787,476632,RNA-Seq,METATRANSCRIPTOMIC,PAIRED,ILLUMINA,Illumina MiSeq,simple,WUHAN UNIVERSITY
,,,,,,,,,,
Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName
SRR10903402,SRS6007143,SAMN13872786,676694,RNA-Seq,METATRANSCRIPTOMIC,PAIRED,ILLUMINA,Illumina MiSeq,simple,WUHAN UNIVERSITY
,,,,,,,,,,
Run,Sample,BioSample,spots,LibraryStrategy,LibrarySource,LibraryLayout,Platform,Model,SampleType,CenterName
# technology
cat sra/metadata.txt | grep -v "^," | grep -v "^Run" | cut -f9 -d','| sort | uniq -c
1 BGISEQ-500
248 GridION
7 Illumina HiSeq 3000
18 Illumina MiSeq
1 Illumina MiniSeq
2 Ion Torrent S5
3 MinION
8 NextSeq 500
# center
cat sra/metadata.txt | grep -v "^," | grep -v "^Run" | cut -f11 -d','| sort | uniq -c
1 "SHANGHAI PUBLIC HEALTH CLINICAL CENTER & SCHOOL OF PUBLIC HEALTH
11 "WUHAN INSTITUTE OF VIROLOGY
1 BGI
2 HKU-SHENZHEN HOSPITAL
1 THE SCRIPPS RESEARCH INSTITUTE
2 UFBA
1 UNIVERSIDAD TECNOLOGICA DE PEREIRA
1 UNIVERSITY OF HONG KONG
3 UNIVERSITY OF MELBOURNE
14 UNIVERSITY OF WASHINGTON
8 UNIVERSITY OF WISCONSIN - MADISON
243 WUHAN UNIVERSITY
cat sra/metadata.txt | grep MELBOURNE
SRR11267570,SRS6201528,SAMN14167851,779208,RNA-Seq,VIRAL RNA,SINGLE,OXFORD_NANOPORE,GridION,simple,UNIVERSITY OF MELBOURNE
SRR11350376,SRS6201528,SAMN14167851,779208,RNA-Seq,VIRAL RNA,SINGLE,OXFORD_NANOPORE,GridION,simple,UNIVERSITY OF MELBOURNE
SRR11300652,SRS6313628,SAMN14371025,430923,RNA-Seq,VIRAL RNA,SINGLE,OXFORD_NANOPORE,GridION,simple,UNIVERSITY OF MELBOURNEDownload all Illumina data for further analysis. (I have not analysed Oxford Nanopore before, so I won't download these yet.)
for acc in `cat sra/metadata.txt | grep ILLUMINA | cut -f1 -d','`; do
echo $acc
fasterq-dump -p --outdir raw/fastq $acc
doneWe will analysis the data from SRR10971381 following the preprocessing steps outlined in https://github.com/galaxyproject/SARS-CoV-2/blob/master/1-PreProcessing/pp_wf.png.
Use SRA Toolkit to download FASTQ sequences from the SRA. First use prefetch, a command-line for downloading SRA, dbGaP, and ADSP data, to download sra files. See https://www.ncbi.nlm.nih.gov/books/NBK242621/ for more information.
Using prefetch kept resulting in timeout errors. Perhaps https://github.com/ncbi/sra-tools/wiki/06.-Connection-Timeouts will help?
prefetch --output-directory raw SRR10971381
# connection keeps dying
wget https://sra-download.ncbi.nlm.nih.gov/traces/sra46/SRR/010714/SRR10971381
md5sum SRR10971381 > SRR10971381.md5sum
cat SRR10971381.md5sum
5496488662893a836e23541b84bfb7cd SRR10971381I have uploaded the SRA object SRR10971381 to my web server: https://davetang.org/file/SRR10971381. You can download it from there.
wget -c -N https://davetang.org/file/SRR10971381
wget -c -N https://davetang.org/file/SRR10971381.md5sum
md5sum -c SRR10971381.md5sum
SRR10971381: OKNext use fastq-dump to convert SRA data into FASTQ.
fastq-dump --split-files ./SRR10971381
2020-03-10T14:51:00 fastq-dump.2.10.3 err: name not found while resolving query within virtual file system module - failed to resolve accession './SRR10971381' - Cannot resolve accession ( 404 )
Read 28282964 spots for ./SRR10971381
Written 28282964 spots for ./SRR10971381Preprocess using fastp.
fastp --thread 8 -i SRR10971381_1.fastq -I SRR10971381_2.fastq -o SRR10971381_1_fastp.fastq -O SRR10971381_2_fastp.fastq
Read1 before filtering:
total reads: 28282964
total bases: 4017125680
Q20 bases: 1783384314(44.3945%)
Q30 bases: 1735659038(43.2065%)
Read2 before filtering:
total reads: 28282964
total bases: 4013917534
Q20 bases: 1723725994(42.9437%)
Q30 bases: 1652844944(41.1779%)
Read1 after filtering:
total reads: 13054241
total bases: 1786633510
Q20 bases: 1671652872(93.5644%)
Q30 bases: 1634552420(91.4878%)
Read2 aftering filtering:
total reads: 13054241
total bases: 1782180210
Q20 bases: 1625652911(91.2171%)
Q30 bases: 1568126467(87.9892%)
Filtering result:
reads passed filter: 26108482
reads failed due to low quality: 30441256
reads failed due to too many N: 16190
reads failed due to too short: 0
reads with adapter trimmed: 582728
bases trimmed due to adapters: 30162896
Duplication rate: 5.57505%
Insert size peak (evaluated by paired-end reads): 150
JSON report: fastp.json
HTML report: fastp.html
fastp --thread 8 -i SRR10971381_1.fastq -I SRR10971381_2.fastq -o SRR10971381_1_fastp.fastq -O SRR10971381_2_fastp.fastq --thread 8
fastp v0.20.0, time used: 544 secondsQuality control using FastQC.
mkdir fastqc_out
fastqc -o fastqc_out -f fastq SRR10971381_1_fastp.fastq SRR10971381_2_fastp.fastqUse BWA MEM to map raw reads back to MN908947.
mkdir bwa_index
cp MN908947.fa bwa_index
cd bwa_index
bwa index MN908947.fa
bwa mem -t 8 raw/bwa_index/MN908947.fa raw/SRR10971381/SRR10971381_1_fastp.fastq raw/SRR10971381/SRR10971381_2_fastp.fastq | samtools sort - -o result/SRR10971381_MN908947.bamStats on the BAM file.
samtools flagstat -@8 SRR10971381_MN908947.bam
26137357 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
28875 + 0 supplementary
0 + 0 duplicates
174256 + 0 mapped (0.67% : N/A)
26108482 + 0 paired in sequencing
13054241 + 0 read1
13054241 + 0 read2
136034 + 0 properly paired (0.52% : N/A)
136414 + 0 with itself and mate mapped
8967 + 0 singletons (0.03% : N/A)
0 + 0 with mate mapped to a different chr
0 + 0 with mate mapped to a different chr (mapQ>=5)
samtools view -F 0x804 -f 2 -b SRR10971381_MN908947.bam > SRR10971381_MN908947_mapped.bambcftools mpileup -f raw/MN908947.fa result/SRR10971381_MN908947_mapped.bam | bcftools call -mv -Ov -o result/SRR10971381_MN908947_mapped.vcf- https://github.com/CSSEGISandData/COVID-19
- https://github.com/galaxyproject/SARS-CoV-2
- https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/
- https://www.sciencemag.org/news/2020/01/mining-coronavirus-genomes-clues-outbreak-s-origins
- https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6356540/
See Entrez Direct (EDirect) for more information.
Navigation functions support exploration within the Entrez databases:
- esearch performs a new Entrez search using terms in indexed fields.
- elink looks up neighbors (within a database) or links (between databases).
- efilter filters or restricts the results of a previous query.
Records can be retrieved in specified formats or as document summaries:
- efetch downloads records or reports in a designated format.
Desired fields from XML results can be extracted without writing a program:
- xtract converts EDirect XML output into a table of data values.
Several additional functions are also provided:
- einfo obtains information on indexed fields in an Entrez database.
- epost uploads unique identifiers (UIDs) or sequence accession numbers.
- nquire sends a URL request to a web page or CGI service.