This repository covers some bioinformatics analysis applied to phosphoproteomics study.
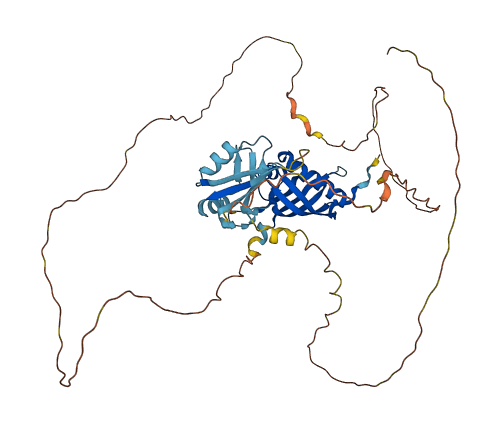
Ras GTPase-activating protein-binding protein 2 (G3BP2) predicted by AlphaFold2. This protein has several ubiquitination sites annotated in PhosphoSitePlus.
- Phosphoproteomics
- Instalation dependencies
- Notebooks
- Useful databases for PTM analysis
- Author
- License
Install dependencies with mamba
mamba create --name phosphoproteomics python=3 pandas matplotlib numpy biopython plotly pypdb- 01_file-processing.ipynb: File processing.
- 02_structural-characterization.ipynb: Structural characterization of phosphorylation sites: analysis of the region, secondary structure and accessibility of the phospho-S/T/Y peptides.
- 03_motif-analysis.ipynb: Phosphorylation motif analysis classification.
- 04_biochemical-properties-analysis.ipynb: Are the phospho-peptides: Acidic-, basic-,proline-,asparagine-, or hydrophobic-directed?
- 05_clustering-analysis.ipynb: PTM clusters analysis.
- 06_motifX.ipynb: Identification of new motifs with MotifX.
- dbPTM: an integrated resource for protein post-translational modifications (PTMs).
- disprot: database of intrinsically disordered proteins.
- MobiDB: a database of protein disorder and mobility annotations.
- Ochoa, D., Jarnuczak, A.F., Viéitez, C. et al. The functional landscape of the human phosphoproteome. Nat Biotechnol 38, 365–373 (2020). https://doi.org/10.1038/s41587-019-0344-3
- Phospho.ELM: a database of S/T/Y phosphorylation sites.
- PhosphoSitePlus: information and tools for the study of protein post-translational modifications (PTMs)
- ProteomeScout: database of proteins and post-translational modifications.
- PTMcode 2: known and predicted functional associations between protein post-translational modifications (PTMs).
- Qoukka: webserver for prediction of kinase family-specific phosphorylation sites.
- Uniprot: comprehensive, high-quality and freely accessible resource of protein sequence and functional information
Fernando Pozo - @fpozoca - Google Scholar – fpozoc@gmx.com - github.com/fpozoc
See LICENSE file.
