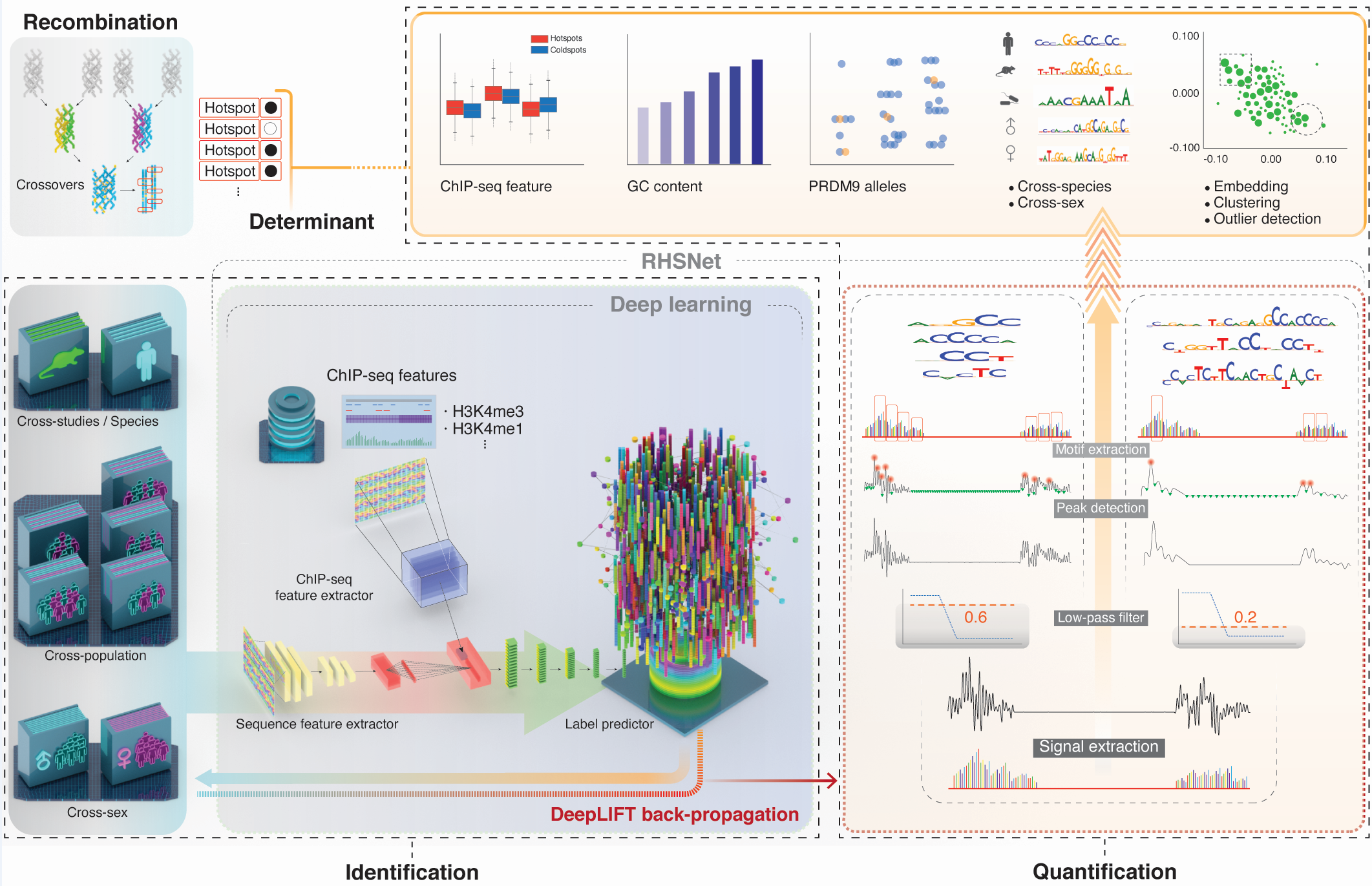
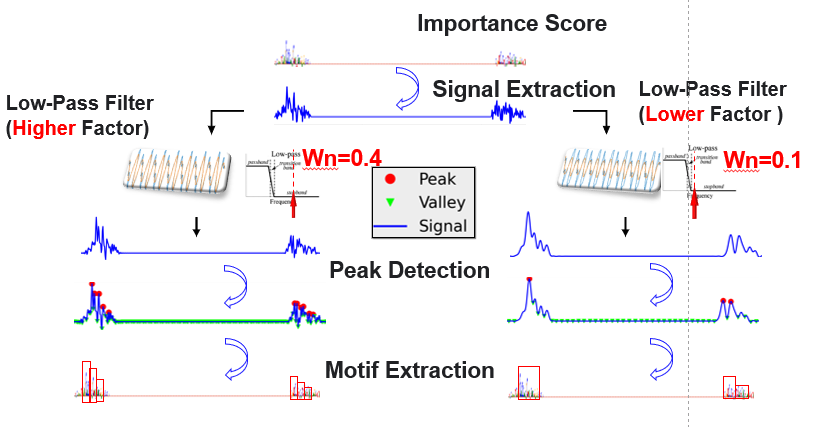
This is the program implementation of the paper: Deep learning identifies and quantifies the recombination hotspot determinants. (currently under submission)
State-of-the-art results are achieved on challenging benchmark datasets across different populations, sexes , and speceis. You can reproduce our results using this repo.
The code is developed using python 3.6 on Ubuntu 16.04. CUDA version is set to be 10.2. NVIDIA GPUs are needed. The code is developed and tested using 2 NVIDIA TITAN GPU cards. Other platforms or GPU cards are not fully tested.
Make sure to create the virtual environment by setting:
conda create -n rhsnet python=3.6
source activate rhsnet
Before running the experiments, pleas make sure that you have the following requirements:
tensorflow-gpu==1.12.0, keras==2.2.4, cudatoolkit==9.0,and cudnn=7.1.2. You can install the dependence by running:
conda install tensorflow-gpu==1.12.0 keras==2.2.4 cudatoolkit==9.0 cudnn=7.1.2 h5py
To install other required packages, please run the following:
pip install -r requirements.txt
The pre-processed dataset for 5-fold cross validation and motif extraction is now publicaly available. You can directly download the dataset from Google Drive
After downloading the dataset, please make sure to put all the folders in the the directory:dataset and make them looks like this:
$ {RHSNet-master}
|-- dataset
├── AFR
├── AMR
├── EAS
├── EUR
├── SAS
├── human_science_2019
├── maternal_science_2019
├── paternal_science_2019
├── nature_2020
├── nature_genetics_2008
├── mouse_cell_2016
To be more specific:
The human_science_2019 repo refers to the Icelandic Human dataset published on Science 2019;
The nature_genetics_2008 repo refers to HapMap II datset;
The nature_2020 repo refers to Sperm dataset published on Nature in Junee 2020;
The maternal_science_2019, paternal_science_2019 repo refers to the generated hotspots and coldspots generated from paternal map and maternal map of the Icelandic Human dataset;
The mouse_cell_2016 repo refers to the mouse dataset published on Cell 2016;
The raw data could be downloaded from Google Drive
For HapMap II or Sperm 2020 dataset, simply replace the directory name from human_science_2019 to nature_genetics_2008 or nature_2020
For Mouse dataset, simply replace the directory name from human_science_2019 to ``mouse_cell_2016
python 5_fold_cross_validation.py experiments/human_science_2019/1000_4/CNN/baseline_classification.json
python 5_fold_cross_validation.py experiments/human_science_2019/1000_4/CNN/mc_rc_classification.json
python 5_fold_cross_validation.py experiments/human_science_2019/1000_4/RHSNet/attention_classification.json
python 5_fold_cross_validation.py experiments/maternal_science_2019/1000_4/RHSNet/reinforce_chip_seq.json
python 5_fold_cross_validation.py experiments/paternal_science_2019/1000_4/RHSNet/reinforce_chip_seq.json
python 5_fold_cross_validation.py experiments/26_population/AFR.json
python 5_fold_cross_validation.py experiments/26_population/AMR.json
python 5_fold_cross_validation.py experiments/26_population/EAS.json
python 5_fold_cross_validation.py experiments/26_population/EUR.json
python 5_fold_cross_validation.py experiments/26_population/SAS.json
The trained model weights are also available in the folder model from Google Drive
python motif_extractor.py experiments/nature_genetics_2008/1000_4/CNN/baseline_classification.json
You can check motif_extractor_example.ipynb for a quick demo on the concept proof.
After running the above command, you should get a dir named /motifs and the following files:
└── nature_genetics_2008
├── filter0.1
├── filter0.1_motifs.json
├── filter0.2
├── filter0.2_motifs.json
├── filter0.4
├── filter0.4_motifs.json
├── recomb_rate.npy
└── scores.npy
If you use our code or models in your research, please cite with:
Li, Y., Chen, S., Rapakoulia, T., Kuwahara, H., Yip, K. Y., & Gao, X. (2021). Deep learning identifies and quantifies recombination hotspot determinants. bioRxiv.