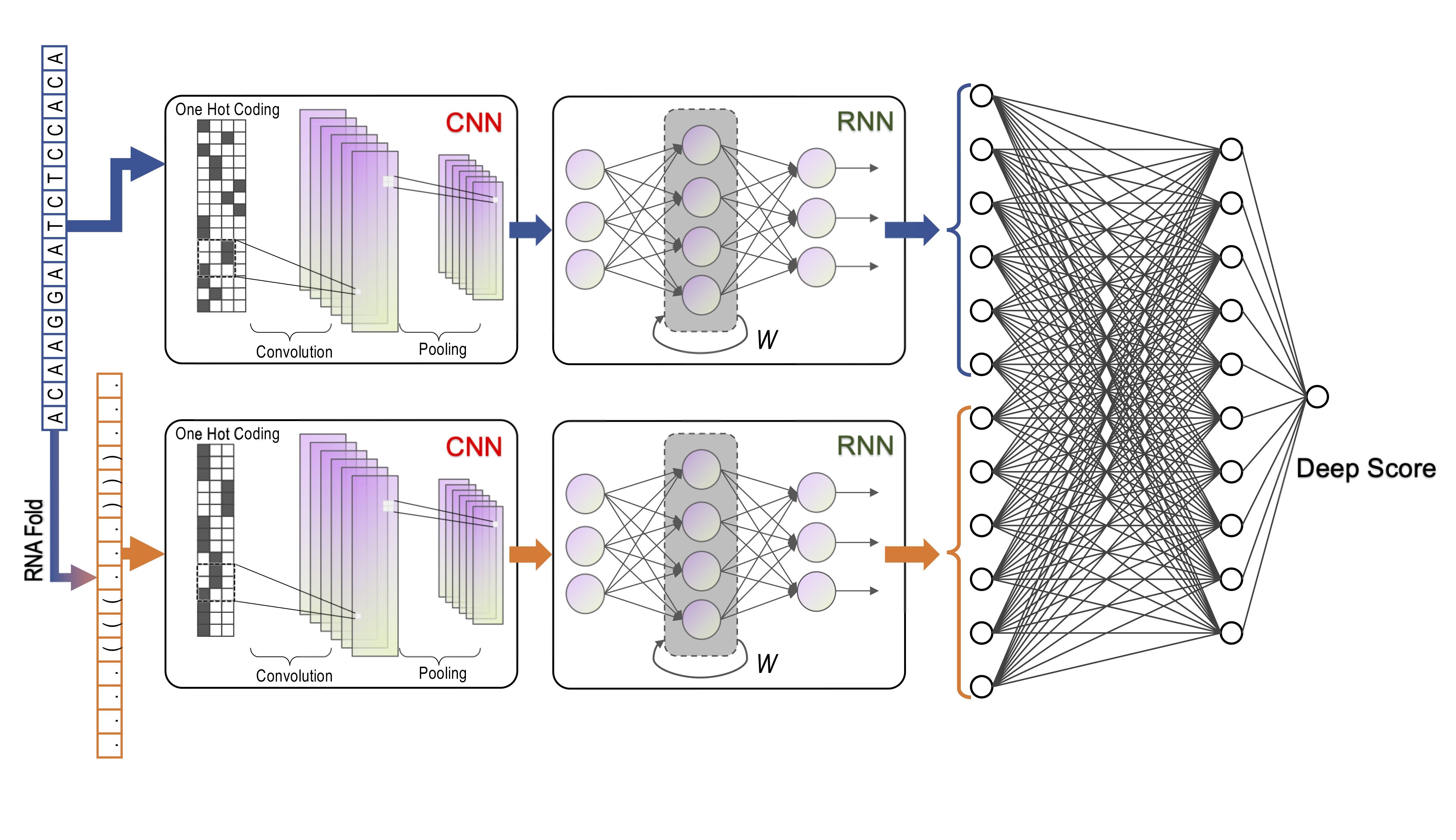
DeepCas13, A deep learning model to predict the CRISPR-Cas13d sgRNA on-target activity with high accuracy from sgRNA sequences and RNA secondary structures.
DeepCas13 uses convolutional recurrent neural network (CRNN) to extract spatial-temporal features for both sequence and secondary structure of a specific sgRNA and then predicts the efficiency by a fully connected neural network (FCNN).
We also provide a webserver for user to design Cas13d sgRNAs online.
- Create a conda environment
conda create --name deepcas13 python=3.6
- Activate the environment
conda activate deepcas13
- Install the dependencies
conda install pandas numpy=1.19.1 seaborn scikit-learn
conda install -c bioconda viennarna
pip install jupyterlab
pip install tensorflow==2.4
Here, we provie 3 demos to show how to use DeepCas13, including train model, predict sgRNA efficiency and design sgRNAs for target sequence.
We can train DeepCas13 model by following command:
python deepcas13.py --train --savepath DL_model --data data/training_data.csv
The input training data is a two-column file, in which the first col is sgRNA sequence and the second col the LFC.
| Parameter | info |
|---|---|
--train |
Set training mode |
--savepath |
Specify the path to save the model |
--data |
The training data |
If the sgRNAs already exist, we can load pretrained model and run DeepCas13 to predict the efficiency:
python deepcas13.py --seq data/test_data_sgrna1.csv --model DL_model
| Parameter | info |
|---|---|
--seq |
The sequence input |
--model |
Specify the path to the pretrained model |
To design sgRNA for a specific target sequence, DeepCas13 can identify all possible sgRNAs and predict the efficiency:
python deepcas13.py --seq data/test_data_target.fa --model DL_model --type target --length 22
| Parameter | info |
|---|---|
--seq |
The sequence input |
--model |
Specify the path to the pretrained model |
--length |
The sgRNA length |
--type |
The acceptable prediction type: 1) sgrna (default): predict the on-target efficiency of sgRNAs; 2) target: design sgRNAs for the input target sequence |
The output of DeepCas13 is a csv file:
| sgrna | seq | deepscore |
|---|---|---|
| sgRNA_0_22 | TTCCCTACTTCCTGTGCTCTTG | 0.44 |
| sgRNA_1_23 | TCCCTACTTCCTGTGCTCTTGC | 0.61 |
| sgRNA_2_24 | CCCTACTTCCTGTGCTCTTGCG | 0.61 |
| sgRNA_3_25 | CCTACTTCCTGTGCTCTTGCGG | 0.64 |
- Xiaolong Cheng: xcheng@childrensnational.org
- Wei Li: wli2@childrensnational.org
Licensed under the MIT license. This project may not be copied, modified, or distributed except according to those terms.