- A Peptide Feature Extraction tool for Mass Spectrometry Data
- A Logistic Classifier, learning the features of uniprot annotated peptides.
Package Install:
pip install git+ssh://git@github.com/jancr/ppv.git#egg=ppvDeveloper Install:
Clone the PPV repository:
mkdir ppv-project
cd ppv-project
git clone https://github.com/jancr/ppv.gitInstall the package
cd ppv
pip install -e .Download Data:
data from the paper can be found at https://github.com/jancr/ppv-data
Lets go back to the ppv-project folder and clone this repo
cd ..
git clone https://github.com/jancr/ppv-dataThen unzip all the files
cd ppv-data/models
gunzip *.gz
cd ../features
gunzip mouse_features_paper.pickle.gz
cd ../..Hopefully your ppv-project directory now looks like this:
$ ls -lh
drwxrwxr-x 6 jcr jcr 4.0K Mar 3 15:15 ppv
drwxrwxr-x 6 jcr jcr 4.0K Mar 3 15:21 ppv-dataThere are two core file types in this project
UPF files: In the ppv-data/upf there are two types of files. The *.upf:code:
file which contains 1 line per peptide per sample. It had 3 important concepts:
- Meta Data: The field
accnois the sample id to link it to meta data such as "This is Mouse 5" - Peptide ID: the fields
prot_acc,pep_start,pep_stopandpep_mod_seqamounts to the peptide ID, thepep_mod_seqallows us to have seperate ID's for peptides with different PTMs - Abundance: the field
intensityis the abundance recorded by the Mass Spectrometer.
Sample Meta files: These files contain meta data about the upf file, this
is necessary for defining groups when doing statistical analysis of the data,
in relation to the PPV algorithm the only field that matters is rs_acc
which is used to link to the accno field in the upf file, and
subject which is the mouse id.
If you want to use the algorithm for your own data you have to convert the output from the MS into this format.
There are two use cases for this project
- use our model to make predictions for your own data
- train your own model on your (and our?) data
In either case you need to extract features from your data. Before you can train or predict, so let's do that
All the features can be found in
ppv-data/features/mouse_features_paper.pickle.gz, this file contains
all the features extracted from all the tissue files. In order to understand
how this file was created let's create it for 1 tissue, doing it for all simply
amounts to using a for loop :)
Import statements:
import pandas as pd
import peputils
from peputils.proteome import fasta_to_protein_hash
import ppvThen we link to the files in ppv-data:
upf_file = 'upf/mouse_brain_combined.upf'
meta_file = 'upf/mouse_brain_combined.sample.meta'
campaign_name = "Mouse Brain"
mouse_fasta = "uniprot/10090_uniprot.fasta"
known_file = "uniprot/known.tsv"Then we now create a upf data frame, we do this using data frame method
.peptidomics.load_upf_meta, which is defined in peputils:
df_raw = pd.DataFrame.peptidomics.load_upf_meta(upf_file, meta_file, campaign_name)We then normalize this dataframe such that all the peptides found across all samples sum to the same, to correct for different sample loading.
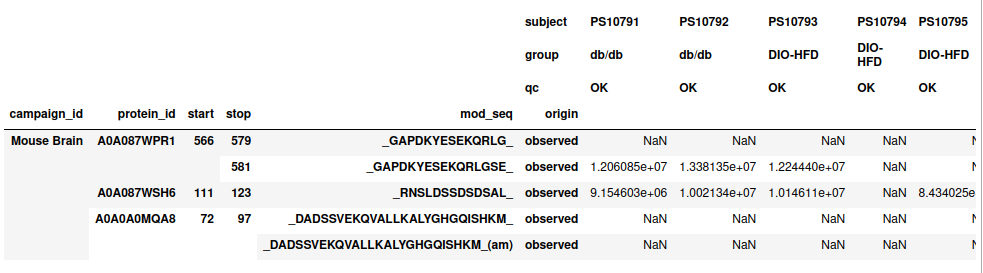
df = df_raw.peptidomics.normalize()Now we have a normalized peptidomics dataframe, it looks like this:
df.head()So much like the .upf file we have 1 row for each observed peptide and 1 column
for each sample abundance.
The above dataframe is what is needed for feature extraction, to extract features from the df use the following method:
n_cpu = 8
mouse_proteins = fasta_to_protein_hash(mouse_fasta)
dataset_features = df.ppv_feature_extractor.create_feature_df(
mouse_proteins, n_cpus=n_cpu, known=known_file, peptides='valid')Note: The feature extraction code is parallelized such that if
n_cpu=8, then it will concurrently extract features from 8 protein backbones,
as some proteins have a much higher number of peptides than others (and the
algorithm scales O(N^2) with the number of peptides in a protein), the progress
bar seem to stall, when there are only the 1-5 proteins with most peptides
left. Be patient my young padowan, the program is not stuck in an infinite
loop, but it may take some hours to finish.
The features from the paper can be loaded from the ppv-data repository:
dataset_features = pd.read_pickle('features/mouse_features_paper.pickle')See section 4 of the next section
The code assumes that the feature generation pipeline was run successfully,
transforming the peptidomics data into a pandas dataframe stored as
mouse_features_paper.pickle. To split the data into 5 folds, run
The script nested_cv.py trains our ML models in nested cross-validation,
yielding 20 models. The script also trains various baseline ML models.
Internally, the PPV model presented in the main papers is called
f_logreg (frequentist logistic regression). If you want to skip
training baseline ML models, comment out the respective models in runs
starting from line 381.
.. code-block:: bash
python3 scripts/nested_cv.py -d ../ppv-data/features/mouse_features_paper_sklearn.pickle -od ../ppv-data/nested_cv
This creates a directory called nested_cv that contains the cross-validated models.
The jupyter notebook notebooks/manuscript_figures.ipynb produces the
performance plots shown in the manuscript from nested_cv and the saved
mouse_features_paper_sklearn.pickle feature data.
the notebooks are saved in markdown format, to convert them to interactive notebook run:
python -m jupytext --to notebook notebooks/manuscript_figures.ipynb
python -m jupytext --to notebook notebooks/plot_validation.ipynbThe full PPV model is an ensemble of the cross-validated models. To apply it to new data, use the following code snippet that takes care of averaging the 20 predictions.
def make_features_numerical(df: pd.DataFrame) -> pd.DataFrame:
'''Cannot train on boolean values.'''
df = df.copy()
for column in df.columns:
if df[column].dtype == bool:
df[column] = df[column].astype(int)
return dfdef predict_probabilities(df, model_dir: str = "nested_cv/cv_f_logreg", folds = [0,1,2,3,4]):
df_X = make_features_numerical(df_X)
X = df_X.values
all_probs = []
# predict from all the test models and average probabilities.
for val in folds:
for test in folds:
if val == test:
continue
model = pickle.load(open(os.path.join(results_dir, f'model_t{test}_v{val}.pkl'), 'rb'))
probs = model.predict_proba(X)[:, 1]
all_probs.append(probs)
probs = np.stack(all_probs).mean(axis=0)
return probs
exclude_features = [
( 'MS Count', 'start'),
( 'MS Count', 'stop'),
( 'MS Frequency', 'protein_coverage'),
( 'MS Frequency', 'cluster_coverage'),
]
feature_columns = df.columns[ (df.columns.get_level_values(0).str.startswith('MS')) & ~(df.columns.isin(exclude_features))]
df['Annotations', 'PPV'] = predict_probabilities(df[feature_columns])