The MCSPACE software package implements a custom, generative AI-based model for identifying, from sequencing data, spatially co-localized groups of microbes, termed assemblages, and detecting changes in the proportions of these assemblages over time and due to introduced perturbations.
git clone https://github.com/gerberlab/MCSPACE.git
pip install MCSPACE/.
Install pytorch from pip
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cpu
pip3 install torch torchvision torchaudio
MCSPACE is implemented as a python library and as a command line interface (CLI). The library can be imported using the command: import mcspace, and the CLI is accessed using the command mcspace. The main classes and methods are documented here.
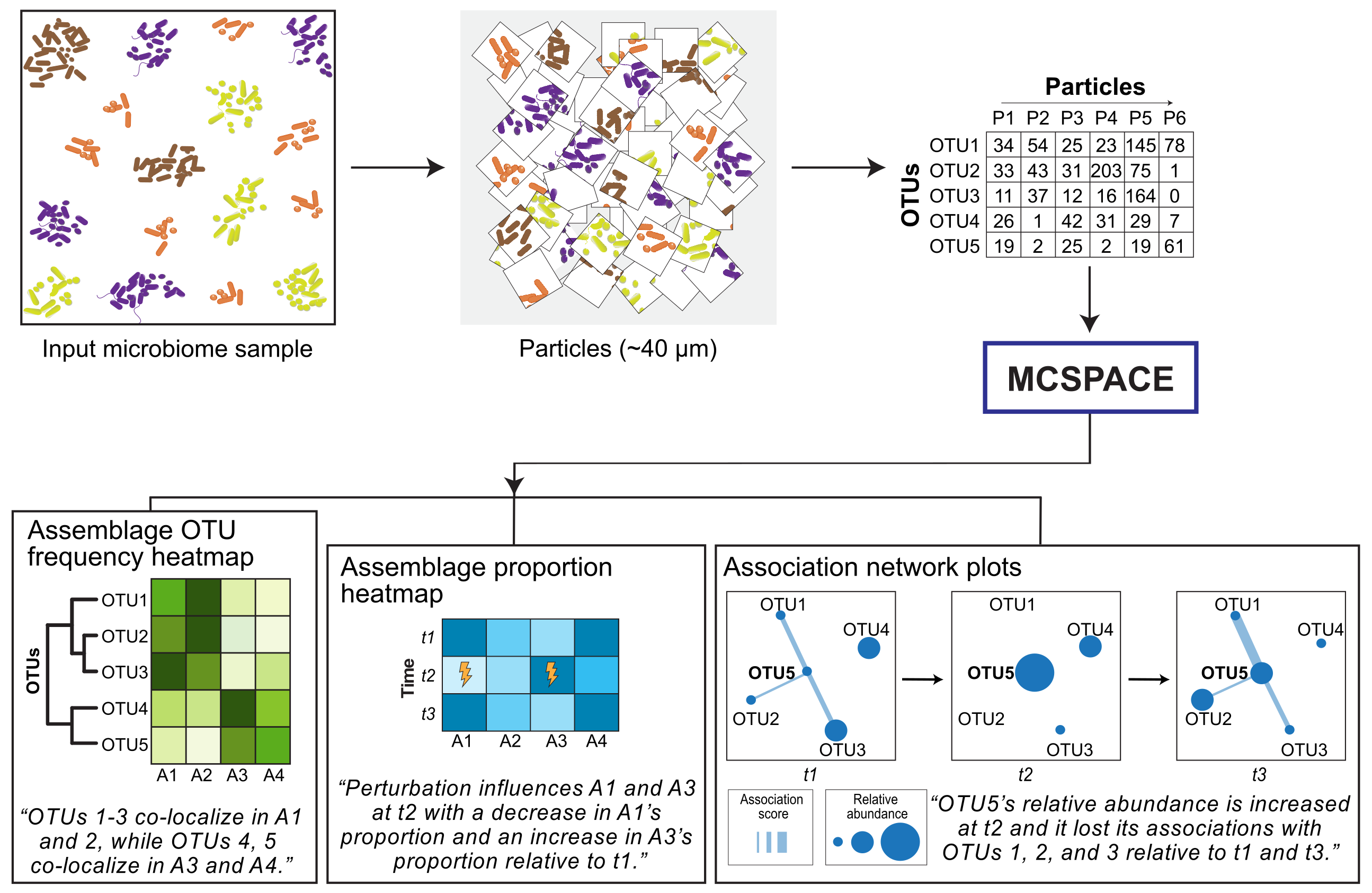 The MCSPACE model takes as input spatial co-localization sequencing data from technologies such as MaPS-seq and SAMPL-seq over time, as well as information on which timepoints correspond to experimental perturbations. The MCSPACE model then infers a sparse set of microbial assemblages and their proportions over time, as well as which assemblages are significantly affected by perturbations. See the tutorials for examples on running the model pipeline.
The MCSPACE model takes as input spatial co-localization sequencing data from technologies such as MaPS-seq and SAMPL-seq over time, as well as information on which timepoints correspond to experimental perturbations. The MCSPACE model then infers a sparse set of microbial assemblages and their proportions over time, as well as which assemblages are significantly affected by perturbations. See the tutorials for examples on running the model pipeline.
To get familiar with the model and software, we recommend going through the provided tutorials here. These go over how to format data for MCSPACE, how to filter and preprocess data for inference, how to run model inference, and methods for visualizing the results.
- Uppal, G., Urtecho, G., Richardson, M., Moody, T., Wang, H.H. and Gerber, G.K., MC-SPACE: Microbial communities from spatially associated counts engine. ICML CompBio.