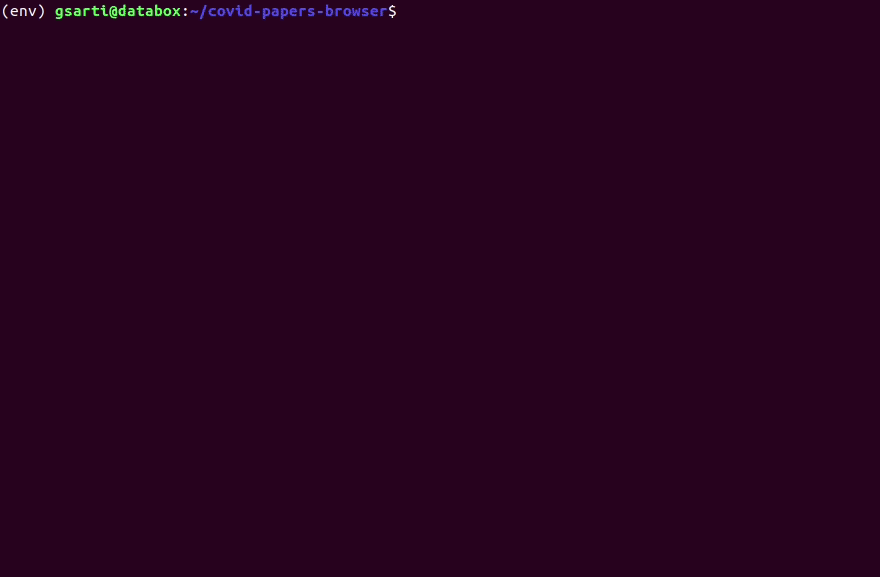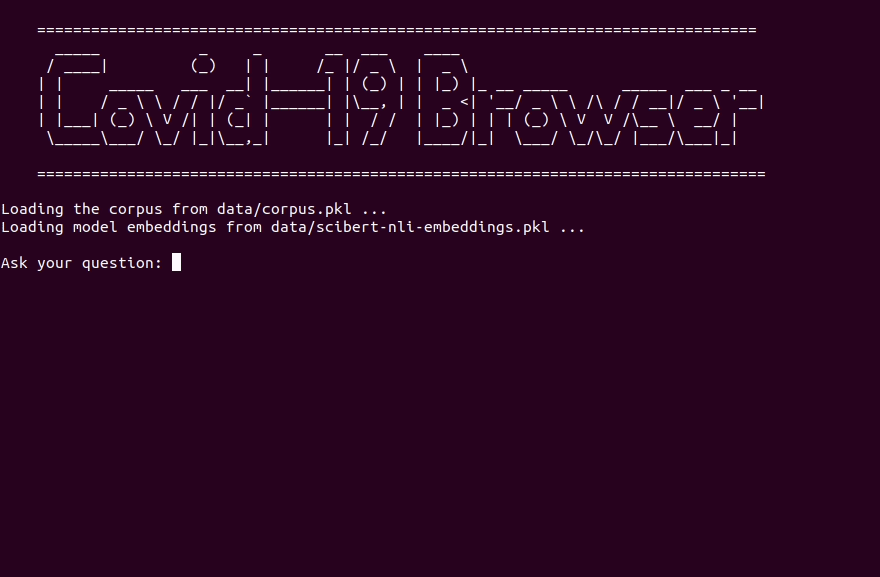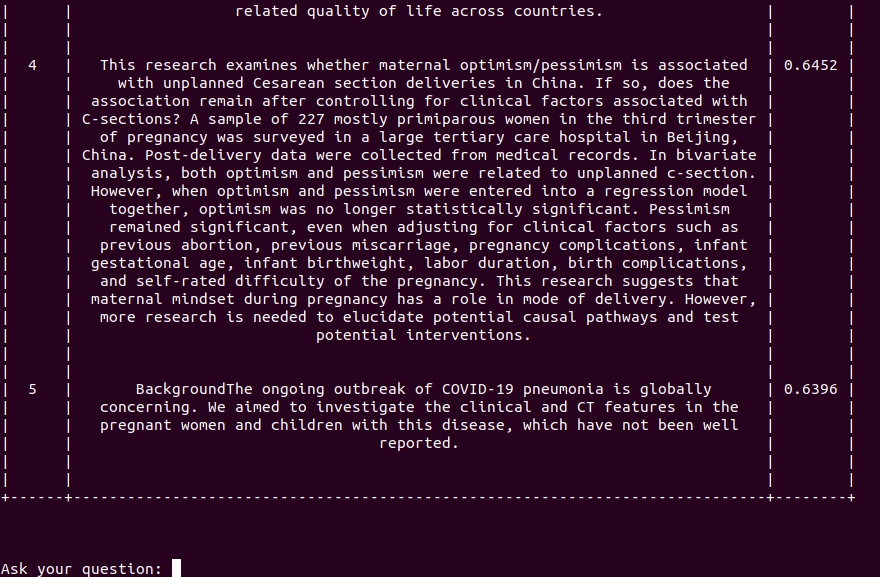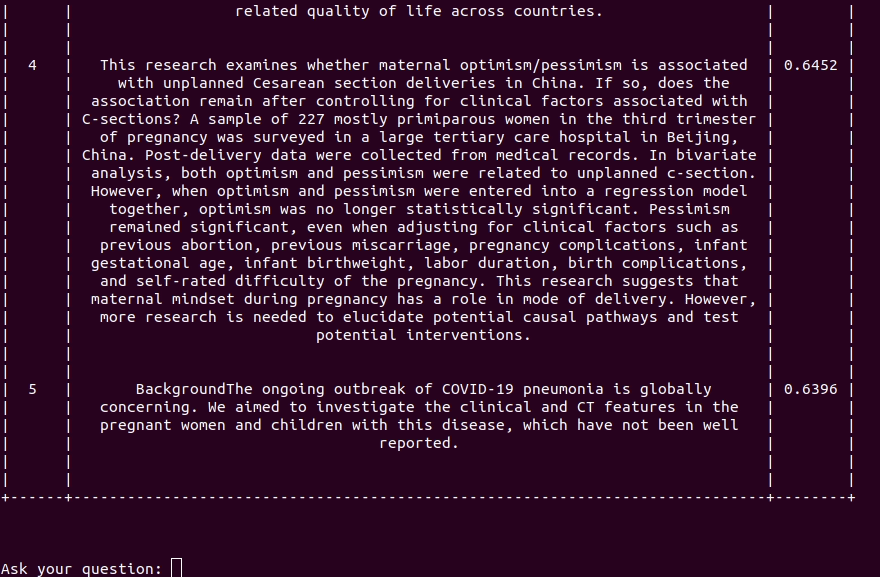
Covid-19 Semantic Browser is an interactive experimental tool leveraging a state-of-the-art language model to search relevant content inside the COVID-19 Open Research Dataset (CORD-19) recently published by the White House and its research partners. The dataset contains over 44,000 scholarly articles about COVID-19, SARS-CoV-2 and related coronaviruses.
Various models already fine-tuned on Natural Language Inference are available to perform the search:
-
scibert-nli, a fine-tuned version of AllenAI's SciBERT [1]. -
biobert-nli, a fine-tuned version of BioBERT by J. Lee et al. [2] -
covidbert-nli, a fine-tuned version of Deepset's CovidBERT. -
clinicalcovidbert-nli, a fine-tuned version of @manueltonneau's ClinicalCovidBERT.
All models are trained on SNLI [3] and MultiNLI [4] using the sentence-transformers library [5] to produce universal sentence embeddings [6]. Embeddings are subsequently used to perform semantic search on CORD-19.
Currently supported operations are:
-
Browse paper abstract with interactive queries.
-
Reproduce SciBERT-NLI, BioBERT-NLI and CovidBERT-NLI training results.
Python 3.6 or higher is required to run the code. First, install the required libraries with pip, then download the en_core_web_sm language pack for spaCy and data for NLTK:
pip install -r requirements.txt
python -m spacy download en_core_web_sm
python -m nltk.downloader punktFirst of all, download a model fine-tuned on NLI from HuggingFace's cloud repository.
python scripts/download_model.py --model scibert-nliSecond, download the data from the Kaggle challenge page and place it in the data folder.
Finally, simply run:
python scripts/interactive_search.pyto enter the interactive demo. Using a GPU is suggested since the creation of the embeddings for the entire corpus might be time-consuming otherwise. Both the corpus and the embeddings are cached on disk after the first execution of the script, and execution is really fast after embeddings are computed.
Use the interactive demo as follows:
First, download a pretrained model from HuggingFace's cloud repository.
python scripts/download_model.py --model scibertSecond, download the NLI datasets used for training and the STS dataset used for testing.
python scripts/get_finetuning_data.pyFinally, run the finetuning script by adjusting the parameters depending on the model you intend to train (default is scibert-nli).
python scripts/finetune_nli.pyThe model will be evaluated against the test portion of the Semantic Text Similarity (STS) benchmark dataset at the end of training. Please refer to my model cards for additional references on parameter values.
[1] Beltagy et al. 2019, "SciBERT: Pretrained Language Model for Scientific Text"
[2] Lee et al. 2020, "BioBERT: a pre-trained biomedical language representation model for biomedical text mining"
[3] Bowman et al. 2015, "A large annotated corpus for learning natural language inference"
[4] Adina et al. 2018, "A Broad-Coverage Challenge Corpus for Sentence Understanding through Inference"
[5] Reimers et al. 2019, "Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks"
[6] As shown in Conneau et al. 2017, "Supervised Learning of Universal Sentence Representations from Natural Language Inference Data"