The main documentation for GSEAPY can be found at https://pythonhosted.org/gseapy
An example to use gseapy, please click here: Example
Release notes : https://github.com/BioNinja/gseapy/releases
GSEAPY could be used for RNA-seq, ChIP-seq, Microarry data. It's used for convenient GO enrichments and produce publishable quality figures in python.
GSEAPY has five sub-commands available: gsea, prerank, ssgsea, replot enrichr.
| gsea: | The gsea module produce GSEA results.The input requries a txt file(FPKM, Expected Counts, TPM, et.al), a cls file, and gene_sets file in gmt format. |
|---|---|
| prerank: | The prerank module produce Prerank tool results. The input expects a pre-ranked gene list dataset with correlation values, which in .rnk format, and gene_sets file in gmt format. prerank module is an API to GSEA pre-rank tools. |
| ssgsea: | The ssgsea module perform single sample GSEA(ssGSEA) analysis. The input expects a gene list with expression values(same with .rnk file, and gene_sets file in gmt format. ssGSEA enrichment score for the gene set as described by D. Barbie et al 2009. |
| replot: | The replot module reproduce GSEA desktop version results. The only input for GSEAPY is the location to GSEA Desktop output results. |
| enrichr: | The enrichr module enable you perform gene set enrichment analysis using Enrichr API. Enrichr is open source and freely available online at: http://amp.pharm.mssm.edu/Enrichr . It runs very fast. |
Please use 'gseapy COMMAND -h' to see the detail description for each option of each module.
The full GSEA is far too extensive to describe here; see
GSEA documentation for more information. All files' formats for GSEApy are identical to GSEA desktop version.
If you use gseapy in your research, you should cite the original ``GSEA`` and ``Enrichr`` paper.
I would like to use Pandas to explore my data, but I did not find a convenient tool to do gene set enrichment analysis in python. So, here is my reason:
- Running inside python interactive console without switch to R!!!
- User friendly for both wet and dry lab usrers.
- Produce or reproduce pubilishable figures.
- Perform batch jobs easy.
- Easy to use in bash shell or your data analysis workflow, e.g. snakemake.
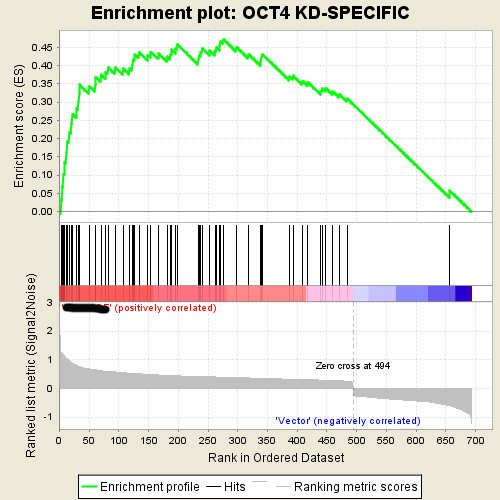
This is an example of GSEA desktop application output
Using the same data from GSEA, GSEAPY reproduce the example above.
Using Prerank or replot module will reproduce the same figure for GSEA Java desktop outputs
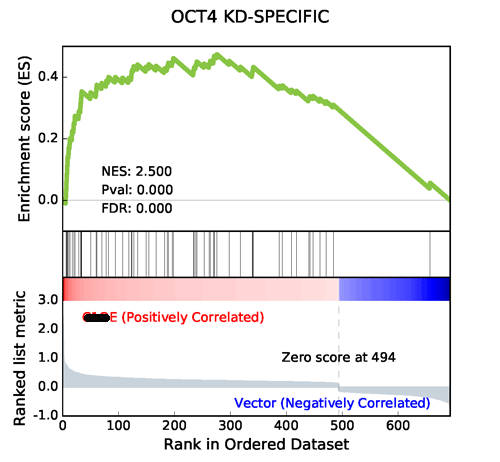
Generated by GSEAPY
GSEAPY figures are supported by all matplotlib figure formats.
You can modify GSEA plots easily in .pdf files. Please Enjoy.
A graphical introduction of Enrichr
The only thing you need to prepeare is a gene list file in txt format(one gene id per row), or a python list object.
Note: Enrichr uses a list of Entrez gene symbols as input. You should convert all gene names to uppercase.
For example, both a list object and txt file are supported for enrchr API
# if you perfer to run gseapy.enrchr() inside python console, you could assign a list object to
# gseapy like this.
gene_list = ['SCARA3', 'LOC100044683', 'CMBL', 'CLIC6', 'IL13RA1', 'TACSTD2', 'DKKL1',
'CSF1', 'CITED1', 'SYNPO2L']# an alternative way is that you could provide a gene list txt file which looks like this:
with open('data/gene_list.txt') as genes:
print(genes.read())
CTLA2B
SCARA3
LOC100044683
CMBL
CLIC6
IL13RA1
TACSTD2
DKKL1
CSF1
CITED1
SYNPO2L
TINAGL1
PTX3# if you have conda
$ conda install -c bioconda gseapy
# for windows users
$ conda install -c bioninja gseapy
# or use pip to install the lastest release
$ pip install gseapy$ pip install git+git://github.com/BioNinja/gseapy.git#egg=gseapy- Python 2.7 or 3.4+
- Numpy
- Pandas
- Matplotlib
- Beautifulsoup4
- Requests(for enrichr API)
You may also need to install lxml, html5lib, if you could not parse xml files.
Unless you know exactly how GSEA works, you should convert all gene symobl names to uppercase first.
# An example to reproduce figures using replot module.
$ gseapy replot -i ./Gsea.reports -o test
# An example to run GSEA using gseapy gsea module
$ gseapy gsea -d exptable.txt -c test.cls -g gene_sets.gmt -o test
# An example to run Prerank using gseapy prerank module
$ gseapy prerank -r gsea_data.rnk -g gene_sets.gmt -o test
# An example to run ssGSEA using gseapy ssgsea module
$ gseapy ssgsea -d expression.txt -g gene_sets.gmt -o test
# An example to use enrichr api
# see details of -g below, -d is optional
$ gseapy enrichr -i gene_list.txt -g KEGG_2016 -d pathway_enrichment -o test- Prepare expression.txt, gene_sets.gmt and test.cls required by GSEA, you could do this
import gseapy
# run GSEA.
gseapy.gsea(data='expression.txt', gene_sets='gene_sets.gmt', cls='test.cls', outdir='test')
# run prerank
gseapy.prerank(rnk='gsea_data.rnk', gene_sets='gene_sets.gmt', outdir='test')
# run ssGSEA
gseapy.ssgsea(data="expression.txt", gene_sets= "gene_sets.gmt", outdir='test')
# An example to reproduce figures using replot module.
gseapy.replot(indir='./Gsea.reports', outdir='test')- If you prefer to use Dataframe, dict, list in interactive python console, you could do this
# assign dataframe, and use enrichr libary data set 'KEGG_2016'
expression_dataframe = pd.DataFrame()
sample_name = ['A','A','A','B','B','B'] # always only two group,any names you like
# assign gene_sets parameter with enrichr library name or gmt file on your local computor.
gseapy.gsea(data=expression_dataframe, gene_sets='KEGG_2016', cls= sample_names, outdir='test')
# using prerank tool
gene_ranked_dataframe = pd.DataFrame()
gseapy.prerank(rnk=gene_ranked_dataframe, gene_sets='KEGG_2016', outdir='test')
# using ssGSEA
gseapy.ssgsea(data=ssGSEA_dataframe, gene_sets='KEGG_2016', outdir='test')- For
enrichr, you could assign a list object or a txt file
# assign a list object to enrichr
l = ['SCARA3', 'LOC100044683', 'CMBL', 'CLIC6', 'IL13RA1', 'TACSTD2', 'DKKL1', 'CSF1',
'SYNPO2L', 'TINAGL1', 'PTX3', 'BGN', 'HERC1', 'EFNA1', 'CIB2', 'PMP22', 'TMEM173']
gseapy.enrichr(gene_list=l, description='pathway', gene_sets='KEGG_2016', outdir='test')
# or a txt file path.
gseapy.enrichr(gene_list='gene_list.txt', description='pathway', gene_sets='KEGG_2016',
outdir='test', cutoff=0.05, format='png' )To see the full list of gseapy supported gene set librarys, please click here: Library
Or use get_library_name function inside python console.
#see full list of latest enrichr library names, which will pass to -g parameter:
names = gseapy.get_library_name()
# show top 20 entries.
print(names[:20])
['Genome_Browser_PWMs',
'TRANSFAC_and_JASPAR_PWMs',
'ChEA_2013',
'Drug_Perturbations_from_GEO_2014',
'ENCODE_TF_ChIP-seq_2014',
'BioCarta_2013',
'Reactome_2013',
'WikiPathways_2013',
'Disease_Signatures_from_GEO_up_2014',
'KEGG_2016',
'TF-LOF_Expression_from_GEO',
'TargetScan_microRNA',
'PPI_Hub_Proteins',
'GO_Molecular_Function_2015',
'GeneSigDB',
'Chromosome_Location',
'Human_Gene_Atlas',
'Mouse_Gene_Atlas',
'GO_Cellular_Component_2015',
'GO_Biological_Process_2015',
'Human_Phenotype_Ontology',]If you would like to report any bugs when you running gseapy, don't hesitate to create an issue on github here, or email me: fangzhuoqing@sibs.ac.cn
Visit the document site at https://pythonhosted.org/gseapy