git clone https://github.com/zhangrengang/SubPhaser
cd SubPhaser
# install
conda env create -f SubPhaser.yaml
conda activate SubPhaser
python setup.py install
# start
cd example_data
# small genome (Arabidopsis_suecica: 270Mb)
bash test_Arabidopsis.sh
# middle genome (peanut: 2.6Gb)
bash test_peanut.sh
# large genome (wheat: 14Gb)
bash test_wheat.sh
For many allopolyploid species, their diploid progenitors are unknown or extinct, making it impossible to unravel their subgenomes.
Here, we develop SubPhaser to partition and phase subgenomes, by using repetitive kmers as the "differential signatures".
The tool also identifies genome-wide subgenome-specific regions and long terminal repeat retrotransposons (LTR-RTs), which will provide insights into the evolutionary history of allopolyploidization.
For details of methods and benchmarking results of SubPhaser, please see the paper in New Phytologist and its Supplementary Material including performances in dozens of chromosome-level neoallopolyploid/hybrid genomes published before October, 2021.
There are mainly four modules:
- The core module to partition and phase subgenomes:
- Count kmers by
jellyfish. - Identify the differential kmers among homoeologous chromosome sets.
- Cluster into subgenomes by a K-Means algorithm and estimate confidence level by the bootstrap.
- Evaluate whether subgenomes are successfully phased by hierarchical clustering and principal component analysis (PCA).
- Count kmers by
- The module to identify and test the enrichments of subgenome-specific kmers:
- Identify subgenome-specific kmers.
- Identify significant enrichments of subgenome-specific kmers by genome window/bin, which is useful to identify homoeologous exchange(s) and/or assembly errors (e.g. switch errors and hamming errors).
- Identify subgenome-specific enrichments with user-defined features (e.g. transposable elements, genes) via
-custom_features.
- The LTR module to identify and analyze subgenome-specific LTR-RT elements (disable by
-disable_ltr):- Identify the LTR-RTs by
LTRharvestand/orLTRfinder(time-consuming for large genome, especiallyLTRfinder). - Classify the LTR-RTs by
TEsorter. - Identify subgenome-specific LTR-RTs by testing the enrichment of subgenome-specific kmers.
- Estimate the insertion age of subgenome-specific LTR-RTs, which is helpful to estimate the time of divergence–hybridization period(s).
- Reconstruct phylogenetic trees of subgenome-specific LTR/Gypsy and LTR/Copia elements, which is helpful to infer the evolutionary history of these LTR-RTs (disable by
-disable_ltrtree, time-consuming for large genome).
- Identify the LTR-RTs by
- The visualization module to visualize genome-wide data (disable by
-disable_circos):- Identify the homoeologous blocks by
minimap2simply (disable by-disable_blocks, time-consuming for large genome). - Integrate and visualize the whole genome-wide data by
circos.
- Identify the homoeologous blocks by
The below is an example of output figures of wheat (ABD, 1n=3x=21):
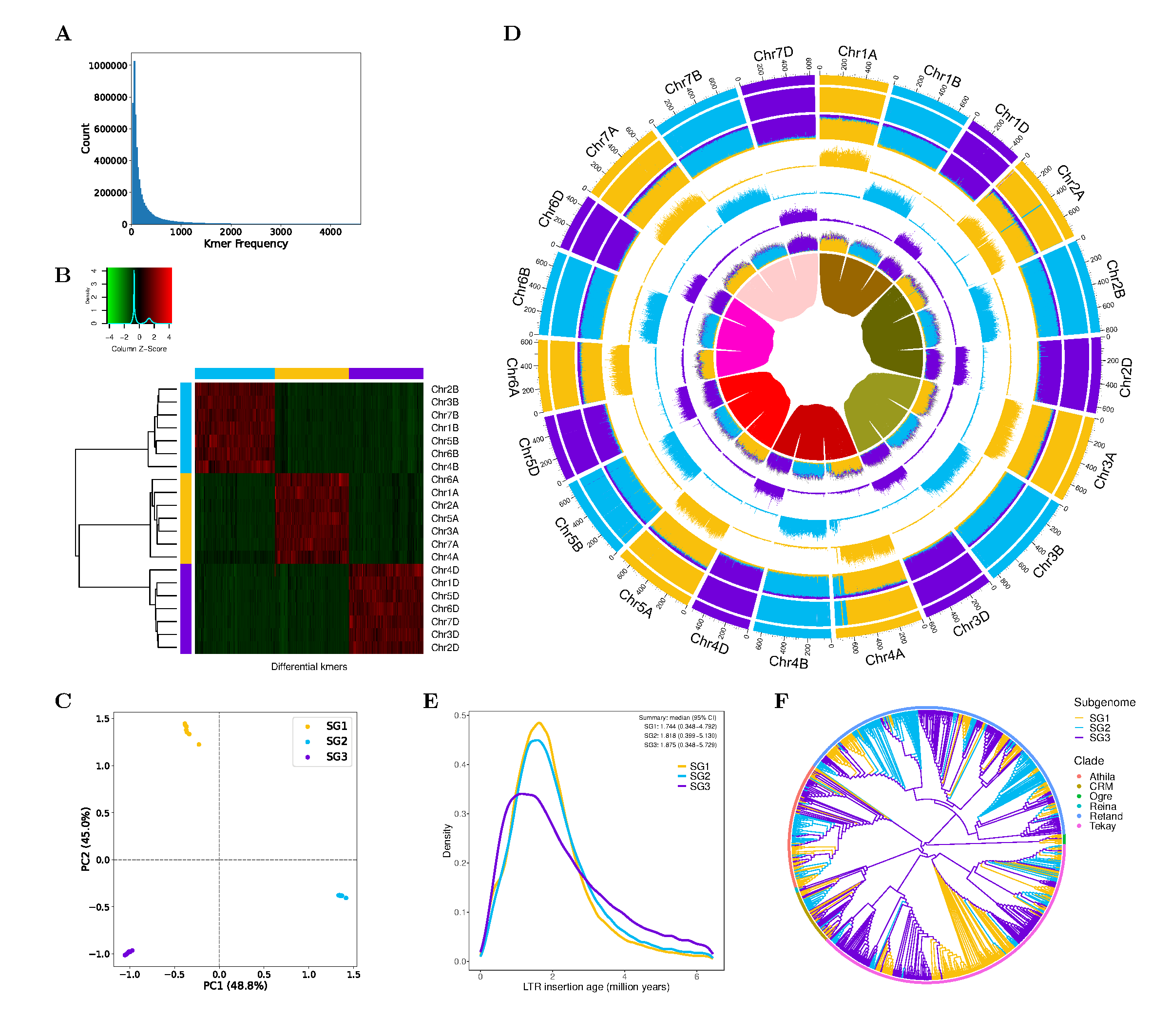 Figure. Phased subgenomes of allohexaploid bread wheat genome. Colors are unified with each subgenome in subplots
Figure. Phased subgenomes of allohexaploid bread wheat genome. Colors are unified with each subgenome in subplots B-F, i.e. the same color means the same subgenome.
- (A) The histogram of differential k-mers among homoeologous chromosome sets.
- (B) Heatmap and clustering of differential k-mers. The x-axis, differential k-mers; y-axis, chromosomes. The vertical color bar, each chromosome is assigned to which subgenome; the horizontal color bar, each k-mer is specific to which subgenome (blank for non-specific kmers).
- (C) Principal component analysis (PCA) of differential k-mers.
- (D) Chromosomal characteristics (window size: 1 Mb). Rings from outer to inner:
- (1) Subgenome assignments by a k-Means algorithm.
- (2) Significant enrichment of subgenome-specific k-mers (blank for non-enriched windows).
- (3) Normalized proportion of subgenome-specific k-mers.
- (4-6) Density distribution (count) of each subgenome-specific k-mer set.
- (7) Density distribution (count) of subgenome-specific LTR-RTs and other LTR-RTs (the most outer, in grey color).
- (8) Homoeologous blocks of each homoeologous chromosome set.
- (E) Insertion time of subgenome-specific LTR-RTs.
- (F) A phylogenetic tree of 1,000 randomly subsampled LTR/Gypsy elements.
- Chromosome-level genome sequences (fasta format), e.g. the wheat genome (ABD, 1n=3x=21).
- Configuration of homoeologous chromosome sets, e.g.
Chr1A Chr1B Chr1D # each row is one homoeologous chromosome set
Chr2B Chr2A Chr2D # chromosome order is arbitrary and useless
Chr3D Chr3B Chr3A # seperate with blank character(s)
Chr4B Chr4D Chr4A
5A|Chr5A 5B|Chr5B 5D|Chr5D # rename chromosome id to 5A, 5B and 5D, respectively
Chr6A,Chr7A Chr6B,Chr7B Chr6D,Chr7D # treat multiple chromosomes together using ","
If some homoeologous relationships are ambiguous, they can be placed as singletons that will not used to identify differential kmers. For example:
Chr1A Chr1B Chr1D
Chr2B Chr2A Chr2D
Chr3D Chr3B Chr3A
Chr4B Chr4D
Chr4A # singleton(s) will skip the step to identify differential kmers
...
- [Optional] Sequences of genomic features (fasta format, with
-custom_features): Any sequences of genomic features, such as transposable elements (TEs), long terminal repeat retrotransposons (LTR-RTs), simple repeats and genes, could be fed to identify the subgenome-specific ones.
Run with default parameters:
subphaser -i genome.fasta.gz -c sg.config
Run with just the core algorithm enabled:
subphaser -i genome.fasta.gz -c sg.config -just_core
or
subphaser -i genome.fasta.gz -c sg.config -disable_ltr -disable_circos
Change key parameters when differential kmers are too few:
subphaser -i genome.fasta.gz -c sg.config -k 13 -q 100 -f 2
Mutiple genomes (e.g. two relative species):
subphaser -i genomeA.fasta.gz genomeB.fasta.gz -c sg.config
Mutiple config files:
subphaser -i genome.fasta.gz -c sg1.config sg2.config
Input custom feature (e.g. transposable element, gene) sequences for subgenome-specific enrichments:
subphaser -i genome.fasta.gz -c sg.config -custom_features TEs.fasta genes.fasta
phase-results/
├── k15_q200_f2.circos/ # config and data files for circos plot, so developers are able to re-plot with some custom modification
├── k15_q200_f2.kmer_freq.pdf # histogram of differential kmers, useful to adjust option `-q`
├── k15_q200_f2.kmer.mat # differential kmer matrix (m kmer × n chromosome)
├── k15_q200_f2.kmer.mat.pdf # heatmap of the kmer matrix
├── k15_q200_f2.kmer.mat.R # R script for the heatmap plot
├── k15_q200_f2.kmer_pca.pdf # PCA plot of the kmer matrix
├── k15_q200_f2.chrom-subgenome.tsv # subgenome assignments and bootstrap values
├── k15_q200_f2.sig.kmer-subgenome.tsv # subgenome-specific kmers
├── k15_q200_f2.bin.enrich # subgenome-specific enrichments by genome window/bin
├── k15_q200_f2.bin.group # grouped bins by potential exchanges based on enrichments
├── k15_q200_f2.ltr.enrich # subgenome-specific LTR-RTs
├── k15_q200_f2.ltr.insert.pdf # density plot of insertion age of subgenome-specific LTR-RTs
├── k15_q200_f2.ltr.insert.R # R script for the density plot
├── k15_q200_f2.LTR_Copia.tree.pdf # phylogenetic tree plot of subgenome-specific LTR/Copia elements
├── k15_q200_f2.LTR_Copia.tree.R # R script for the LTR/Copia tree plot
├── k15_q200_f2.LTR_Gypsy.tree.pdf # phylogenetic tree plot of subgenome-specific LTR/Gypsy elements
├── k15_q200_f2.LTR_Gypsy.tree.R # R script for the LTR/Gypsy tree plot
├── k15_q200_f2.circos.pdf # final circos plot
├── k15_q200_f2.circos.png
├── circos_legend.txt # legend of the circos plot
.....
tmp/
├── LTR.scn # identification of LTR-RTs by LTRharvest and/or LTRfinder
├── LTR.inner.fa # inner sequences of LTR-RTs
├── LTR.inner.fa.cls.* # classfication of LTR-RTs by TEsorter
├── LTR.filtered.LTR.fa # full sequences of the filtered LTR-RTs
├── LTR.LTR_*.aln # alignments of LTR-RTs' protein domains for the below tree
├── LTR.LTR_*.rooted.tre # phylogenetic tree files
├── LTR.LTR_*.map # information of tip nodes on the above tree
.....
If you use SubPhaser, please cite:
Jia K, Wang Z, Wang L et. al. SubPhaser: A robust allopolyploid subgenome phasing method based on subgenome-specific k-mers [J]. New Phytologist, 2022, in press DOI:10.1111/nph.18173
usage: subphaser [-h] -i GENOME [GENOME ...] -c CFGFILE [CFGFILE ...]
[-labels LABEL [LABEL ...]] [-no_label]
[-target FILE] [-sg_assigned FILE] [-sep STR]
[-custom_features FASTA [FASTA ...]] [-pre STR]
[-o DIR] [-tmpdir DIR] [-k INT] [-f FLOAT] [-q INT]
[-baseline BASELINE] [-lower_count INT]
[-min_prop FLOAT] [-max_freq INT] [-max_prop FLOAT]
[-low_mem] [-by_count] [-re_filter] [-nsg INT]
[-replicates INT] [-jackknife FLOAT]
[-max_pval FLOAT]
[-test_method {ttest_ind,kruskal,wilcoxon,mannwhitneyu}]
[-figfmt {pdf,png}]
[-heatmap_colors COLOR [COLOR ...]]
[-heatmap_options STR] [-just_core] [-disable_ltr]
[-ltr_detectors {ltr_finder,ltr_harvest} [{ltr_finder,ltr_harvest} ...]]
[-ltr_finder_options STR] [-ltr_harvest_options STR]
[-tesorter_options STR] [-all_ltr] [-intact_ltr]
[-exclude_exchanges] [-shared_ltr] [-mu FLOAT]
[-disable_ltrtree] [-subsample INT]
[-ltr_domains {GAG,PROT,INT,RT,RH,AP,RNaseH} [{GAG,PROT,INT,RT,RH,AP,RNaseH} ...]]
[-trimal_options STR]
[-tree_method {iqtree,FastTree}] [-tree_options STR]
[-ggtree_options STR] [-disable_circos]
[-window_size INT] [-disable_blocks] [-aligner PROG]
[-aligner_options STR] [-min_block INT]
[-alt_cfgs CFGFILE [CFGFILE ...]] [-chr_ordered FILE]
[-p INT] [-max_memory MEM] [-cleanup] [-overwrite]
[-v]
Phase and visualize subgenomes of an allopolyploid or hybrid based on the repetitive kmers.
optional arguments:
-h, --help show this help message and exit
Input:
Input genome and config files
-i GENOME [GENOME ...], -genomes GENOME [GENOME ...]
Input genome sequences in fasta format [required]
-c CFGFILE [CFGFILE ...], -sg_cfgs CFGFILE [CFGFILE ...]
Subgenomes config file (one homologous group per
line); this chromosome set is for identifying
differential kmers [required]
-labels LABEL [LABEL ...]
For multiple genomes, provide prefix labels for each
genome sequence to avoid conficts among chromosome id
[default: '1-, 2-, ..., n-']
-no_label Do not use default prefix labels for genome sequences
as there is no confict among chromosome id
[default=False]
-target FILE Target chromosomes to output; id mapping is allowed;
this chromosome set is for cluster and phase [default:
the same chromosome set as `-sg_cfgs`]
-sg_assigned FILE Provide subgenome assignments to skip k-means
clustering and to identify subgenome-specific features
[default=None]
-sep STR Seperator for chromosome ID [default="|"]
-custom_features FASTA [FASTA ...]
Custom features in fasta format to enrich subgenome-
specific kmers, such as TE and gene [default: None]
Output:
-pre STR, -prefix STR
Prefix for output [default=None]
-o DIR, -outdir DIR Output directory [default=phase-results]
-tmpdir DIR Temporary directory [default=tmp]
Kmer:
Options to count and filter kmers
-k INT Length of kmer [default=15]
-f FLOAT, -min_fold FLOAT
Minimum fold [default=2]
-q INT, -min_freq INT
Minimum total count for each kmer; will not work if
`-min_prop` is specified [default=200]
-baseline BASELINE Use sub-maximum (1) or minimum (-1) as the baseline of
fold [default=1]
-lower_count INT Don't output k-mer with count < lower-count
[default=3]
-min_prop FLOAT Minimum total proportion (< 1) for each kmer
[default=None]
-max_freq INT Maximum total count for each kmer; will not work if
`-max_prop` is specified [default=1000000000.0]
-max_prop FLOAT Maximum total proportion (< 1) for each kmer
[default=None]
-low_mem Low MEMory but slower [default: True if genome size >
3G, else False]
-by_count Calculate fold by count instead of by proportion
[default=False]
-re_filter Re-filter with subset of chromosomes (subgenome
assignments are expected to change) [default=False]
Cluster:
Options for clustering to phase
-nsg INT Number of subgenomes (>1) [default: auto]
-replicates INT Number of replicates for bootstrap [default=1000]
-jackknife FLOAT Percent of kmers to resample for each bootstrap
[default=50]
-max_pval FLOAT Maximum P value for all hypothesis tests
[default=0.05]
-test_method {ttest_ind,kruskal,wilcoxon,mannwhitneyu}
The test method to identify differiential
kmers[default=ttest_ind]
-figfmt {pdf,png} Format of figures [default=pdf]
-heatmap_colors COLOR [COLOR ...]
Color panel (2 or 3 colors) for heatmap plot [default:
('green', 'black', 'red')]
-heatmap_options STR Options for heatmap plot (see more in R shell with
`?heatmap.2` of `gplots` package) [default="Rowv=T,Col
v=T,scale='col',dendrogram='row',labCol=F,trace='none'
,key=T,key.title=NA,density.info='density',main=NA,xla
b='Differential kmers',margins=c(2.5,12)"]
-just_core Exit after the core phasing module
[default=False]
LTR:
Options for LTR analyses
-disable_ltr Disable this step (this step is time-consuming for
large genome) [default=False]
-ltr_detectors {ltr_finder,ltr_harvest} [{ltr_finder,ltr_harvest} ...]
Programs to detect LTR-RTs [default=['ltr_harvest']]
-ltr_finder_options STR
Options for `ltr_finder` to identify LTR-RTs (see more
with `ltr_finder -h`) [default="-w 2 -D 15000 -d 1000
-L 7000 -l 100 -p 20 -C -M 0.8"]
-ltr_harvest_options STR
Options for `gt ltrharvest` to identify LTR-RTs (see
more with `gt ltrharvest -help`) [default="-seqids yes
-similar 80 -vic 10 -seed 20 -minlenltr 100 -maxlenltr
7000 -mintsd 4 -maxtsd 6"]
-tesorter_options STR
Options for `TEsorter` to classify LTR-RTs (see more
with `TEsorter -h`) [default="-db rexdb -dp2"]
-all_ltr Use all LTR-RTs identified by `-ltr_detectors` (more
LTR-RTs but slower) [default: only use LTR as
classified by `TEsorter`]
-intact_ltr Use completed LTR-RTs classified by `TEsorter` (less
LTR-RTs but faster) [default: the same as `-all_ltr`]
-exclude_exchanges Exclude potential exchanged LTRs for insertion age
estimation and phylogenetic trees [default=False]
-shared_ltr Identify shared LTR-RTs among subgenomes
(experimental) [default=False]
-mu FLOAT Substitution rate per year in the intergenic region,
for estimating age of LTR insertion [default=1.3e-08]
-disable_ltrtree Disable subgenome-specific LTR tree (this step is
time-consuming when subgenome-specific LTR-RTs are too
many, so `-subsample` is enabled by defualt)
[default=False]
-subsample INT Subsample LTR-RTs to avoid too many to construct a
tree [default=1000] (0 to disable)
-ltr_domains {GAG,PROT,INT,RT,RH,AP,RNaseH} [{GAG,PROT,INT,RT,RH,AP,RNaseH} ...]
Domains for LTR tree (Note: for domains identified by
`TEsorter`, PROT (rexdb) = AP (gydb), RH (rexdb) =
RNaseH (gydb)) [default: ['INT', 'RT', 'RH']]
-trimal_options STR Options for `trimal` to trim alignment (see more with
`trimal -h`) [default="-automated1"]
-tree_method {iqtree,FastTree}
Programs to construct phylogenetic trees
[default=FastTree]
-tree_options STR Options for `-tree_method` to construct phylogenetic
trees (see more with `iqtree -h` or `FastTree
-expert`) [default=""]
-ggtree_options STR Options for `ggtree` to show phylogenetic trees (see
more from `https://yulab-smu.top/treedata-book`)
[default="branch.length='none', layout='circular'"]
Circos:
Options for circos plot
-disable_circos Disable this step [default=False]
-window_size INT Window size (bp) for circos plot [default=1000000]
-disable_blocks Disable to plot homologous blocks [default=False]
-aligner PROG Programs to identify homologous blocks
[default=minimap2]
-aligner_options STR Options for `-aligner` to align chromosome sequences
[default="-x asm20 -n 10"]
-min_block INT Minimum block size (bp) to show [default=100000]
-alt_cfgs CFGFILE [CFGFILE ...]
An alternative config file for identifying homologous
blocks [default=None]
-chr_ordered FILE Provide a chromosome order to plot circos
[default=None]
Other options:
-p INT, -ncpu INT Maximum number of processors to use [default=32]
-max_memory MEM Maximum memory to use where limiting can be enabled.
[default=65.2G]
-cleanup Remove the temporary directory [default=False]
-overwrite Overwrite even if check point files existed
[default=False]
-v, -version show program's version number and exit