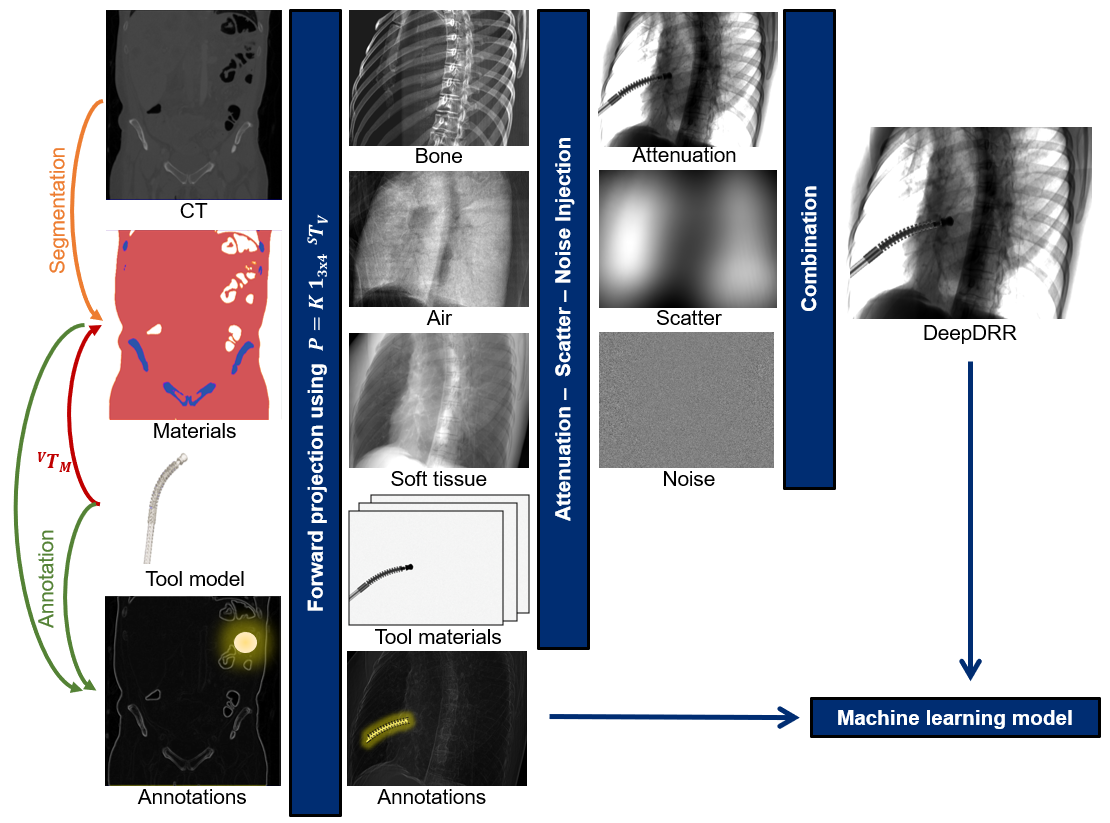
DeepDRR provides state-of-the-art tools to generate realistic radiographs and fluoroscopy from 3D CTs on a training set scale.
DeepDRR requires an NVIDIA GPU, preferably with >11 GB of memory.
- Install CUDA. Version 11 is recommended, but DeepDRR has been used with 8.0
- Make sure your C compiler is on the path. DeepDRR has been used with
gcc 9.3.0 - We recommend installing pycuda separately, as it may need to be built. If you are using Anaconda, run
conda install -c conda-forge pycudato install it in your environment.
- You may also wish to install PyTorch separately, depending on your setup.
- Install from
PyPI
pip install deepdrrInstalling from the dev branch is risky, as it is unstable. However, this installation method can be used for the main branch as well, perhaps somewhat more reliably.
Dependencies:
- CUDA 11.1
- Anaconda
The dev branch contains the most up-to-date code and can be easily installed using Anaconda. To create an environment with DeepDRR, run
git clone https://github.com/arcadelab/deepdrr.git
cd deepdrr
git checkout dev
conda env create -f environment.yaml
conda activate deepdrrDocumentation is available at deepdrr.readthedocs.io.
To create the autodocs, run
sphinx-apidoc -f -o docs/source deepdrrin the base directory. Then do cd docs and make html to build the static site locally.
The following minimal example loads a CT volume from a NifTi .nii.gz file and simulates an X-ray projection:
from deepdrr import geo, Volume, MobileCArm
from deepdrr.projector import Projector # separate import for CUDA init
carm = MobileCArm()
ct = Volume.from_nifti('/path/to/ct_image.nii.gz')
# Initialize the Projector object (allocates GPU memory)
with Projector(ct, carm=carm) as projector:
# Orient and position the patient model in world space.
ct.orient_patient(head_first=True, supine=True)
ct.place_center(carm.isocenter_in_world)
# Move the C-arm to the desired pose.
carm.move_to(alpha=30, beta=10, degrees=True)
# Run projection
image = projector()The script example_projector.py gives an alternative example. Additional tutorials are in progress at deepdrr.readthedocs.io. Contributions are welcome.
Contributions for bug fixes, enhancements, and other suggestions are welcome. Please make a pull request.
DeepDRR combines machine learning models for material decomposition and scatter estimation in 3D and 2D, respectively, with analytic models for projection, attenuation, and noise injection to achieve the required performance. The pipeline is illustrated below.
Further details can be found in our MICCAI 2018 paper "DeepDRR: A Catalyst for Machine Learning in Fluoroscopy-guided Procedures" and the subsequent Invited Journal Article in the IJCARS Special Issue of MICCAI "Enabling Machine Learning in X-ray-based Procedures via Realistic Simulation of Image Formation". The conference preprint can be accessed on arXiv here: https://arxiv.org/abs/1803.08606.
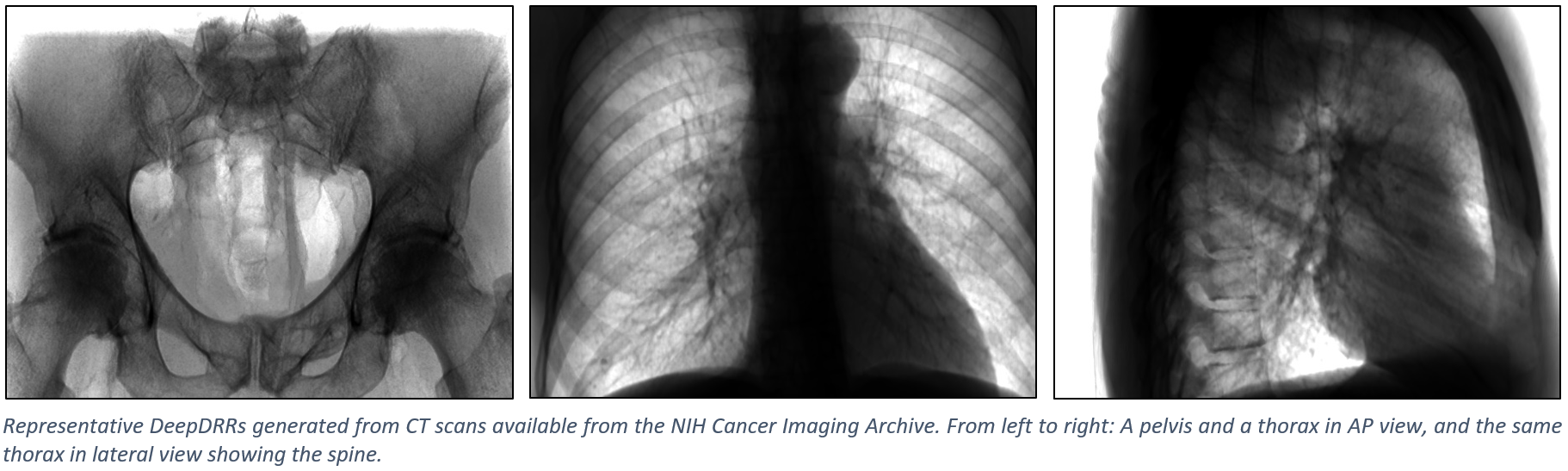
The figure below shows representative radiographs generated using DeepDRR from CT data downloaded from the NIH Cancer Imaging Archive. Please find qualitative results in the Applications section.
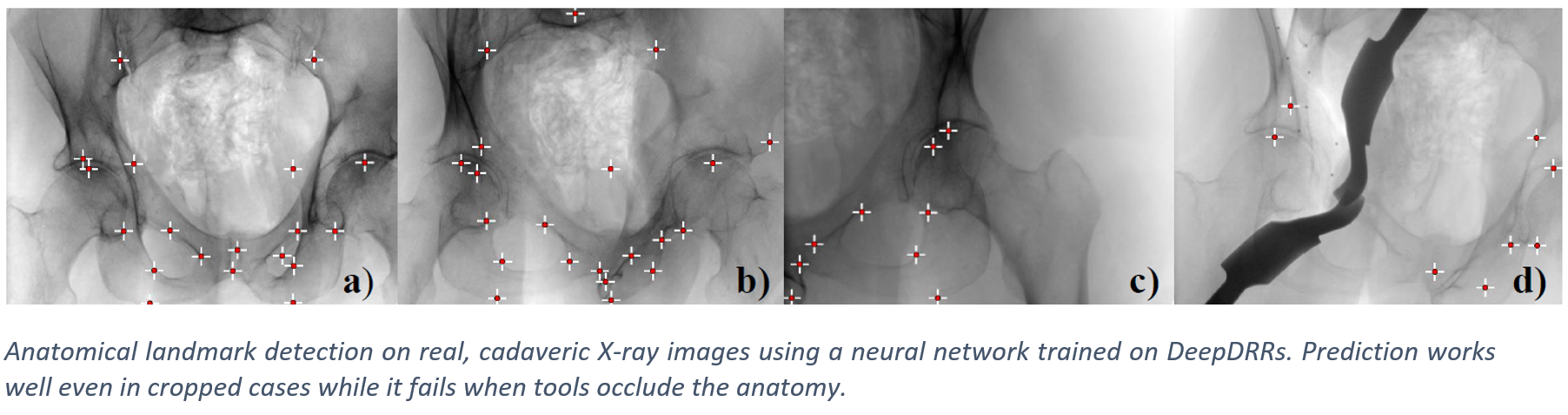
We have applied DeepDRR to anatomical landmark detection in pelvic X-ray: "X-ray-transform Invariant Anatomical Landmark Detection for Pelvic Trauma Surgery", also early-accepted at MICCAI'18: https://arxiv.org/abs/1803.08608 and now with quantitative evaluation in the IJCARS Special Issue on MICCAI'18: https://link.springer.com/article/10.1007/s11548-019-01975-5. The ConvNet for prediction was trained on DeepDRRs of 18 CT scans of the NIH Cancer Imaging Archive and then applied to ex vivo data acquired with a Siemens Cios Fusion C-arm machine equipped with a flat panel detector (Siemens Healthineers, Forchheim, Germany). Some representative results on the ex vivo data are shown below.
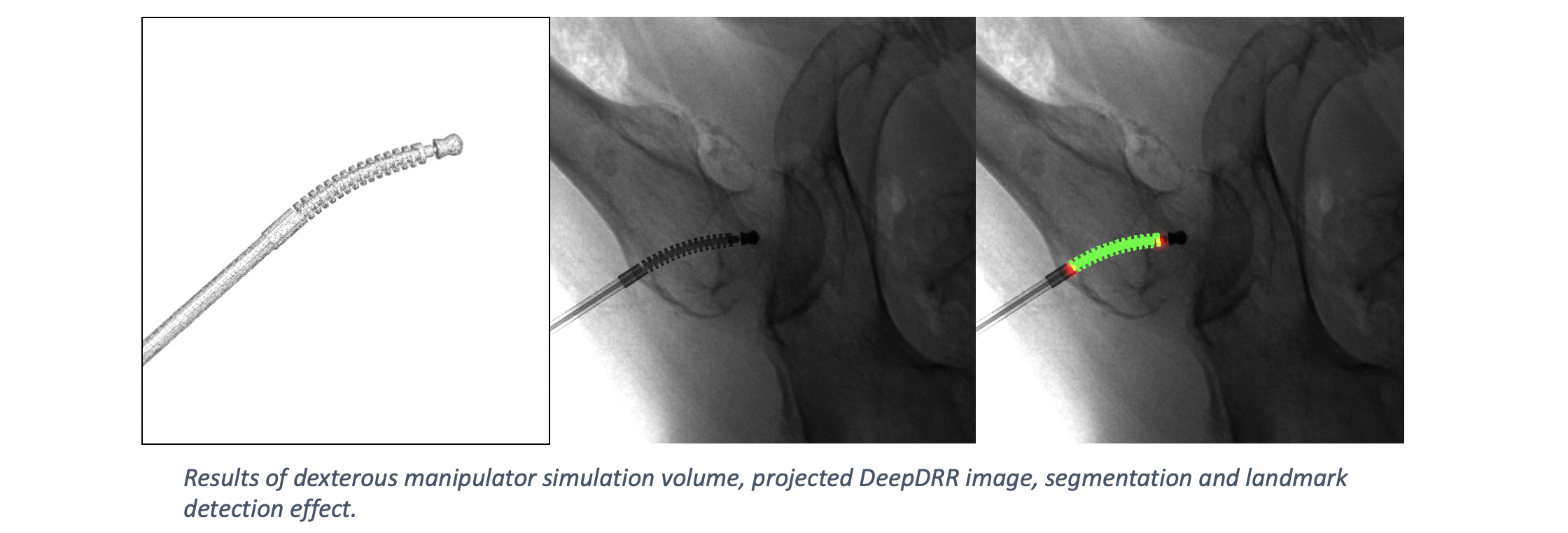
DeepDRR has also been applied to simulate X-rays of the femur during insertion of dexterous manipulaters in orthopedic surgery: "Localizing dexterous surgical tools in X-ray for image-based navigation", which has been accepted at IPCAI'19: https://arxiv.org/abs/1901.06672. Simulated images are used to train a concurrent segmentation and localization network for tool detection. We found consistent performance on both synthetic and real X-rays of ex vivo specimens. The tool model, simulation image and detection results are shown below.
This capability has not been tested in version 1.0. For tool insertion, we recommend working with Version 0.1 for the time being.
- Our material decomposition V-net was trained on NIH Cancer Imagign Archive data. In case it does not generalize perfectly to other acquisitions, the use of intensity thresholds (as is done in conventional Monte Carlo) is still supported. In this case, however, thresholds will likely need to be selected on a per-dataset, or worse, on a per-region basis since bone density can vary considerably.
- Scatter estimation is currently limited to Rayleigh scatter and we are working on improving this. Scatter estimation was trained on images with 1240x960 pixels with 0.301 mm. The scatter signal is a composite of Rayleigh, Compton, and multi-path scattering. While all scatter sources produce low frequency signals, Compton and multi-path are more blurred compared to Rayleigh, suggesting that simple scatter reduction techniques may do an acceptable job. In most clinical products, scatter reduction is applied as pre-processing before the image is displayed and accessible. Consequently, the current shortcoming of not providing full scatter estimation is likely not critical for many applications, in fact, scatter can even be turned off completely. We would like to refer to the Applications section above for some preliminary evidence supporting this reasoning.
- Due to the nature of volumetric image processing, DeepDRR consumes a lot of GPU memory. We have successfully tested on 12 GB of GPU memory but cannot tell about 8 GB at the moment. The bottleneck is volumetric segmentation, which can be turned off and replaced by thresholds (see 1.).
- We currently provide the X-ray source sprectra from MC-GPU that are fairly standard. Additional spectra can be implemented in spectrum_generator.py.
- The current detector reading is the average energy deposited by a single photon in a pixel. If you are interested in modeling photon counting or energy resolving detectors, then you may want to take a look at
mass_attenuation(_gpu).pyto implement your detector. - Currently we do not support import of full projection matrices. But you will need to define K, R, and T seperately or use camera.py to define projection geometry online.
- It is important to check proper import of CT volumes. We have tried to account for many variations (HU scale offsets, slice order, origin, file extensions) but one can never be sure enough, so please double check for your files.
- Currently, the tool/implant model must be represented as a binary 3D volume, rather than a CAD surface model. However, this 3D volume can be of different resolution than the CT volume; particularly, it can be much higher to preserve fine structures of the tool/implant.
- The density of the tool needs to be provided via hard coding in the file 'load_dicom_tool.py' (line 127). The pose of the tool/implant with respect to the CT volume requires manual setup. We provide one example origin setting at line 23-24.
- The tool/implant will supersede the anatomy defined by the CT volume intensities. To this end, we sample the CT materials and densities at the location of the tool in the tool volume, and subtract them from the anatomy forward projections in detector domain (to enable different resolutions of CT and tool volume). Further information can be found in the IJCARS article.
Some issues may arise when using DeepDRR at the same time as PyTorch due to conflicts between pycuda's CUDA initialization and PyTorch CUDA initialization. The best workaround we know of is to first initialize the PyCUDA context (by importing deepdrr.projector) and then run your model on a dummy batch before creating a Projector object. For mysterious reasons (likely involving overlapping GPU resources and the retrograde of Mercury), this seems to work.
import torch
from torch import nn
from torchvision import models
import deepdrr
from deepdrr.projector import Projector # initializes PyCUDA
# Before creating a Projector, run backprop to initialize PyTorch
criterion = nn.CrossEntropyLoss()
model = models.resnet50() # Your model here
model.cuda()
optimizer = optim.SGD(model.parameters(), lr=0.001, momentum=0.9)
optimizer.zero_grad()
x = torch.ones((32, 3, 224, 224), dtype=torch.float32).cuda() # Your image size
y = torch.ones(32, dtype=torch.int64).cuda()
y_pred = model(x)
loss = criterion(y_pred, y)
loss.backward()
optimizer.step()
log.info(f"Ran dummy batch to initialize torch.")
volume = ...
carm = ...
with Projector(volume, carm=carm):
image = projector()
image = image.unsqueeze(0) # add batch dim
y_pred = model(image)
...We hope this proves useful for medical imaging research. If you use our work, we would kindly ask you to reference our work. The MICCAI article covers the basic DeepDRR pipeline and task-based evaluation:
@inproceedings{DeepDRR2018,
author = {Unberath, Mathias and Zaech, Jan-Nico and Lee, Sing Chun and Bier, Bastian and Fotouhi, Javad and Armand, Mehran and Navab, Nassir},
title = {{DeepDRR--A Catalyst for Machine Learning in Fluoroscopy-guided Procedures}},
date = {2018},
booktitle = {Proc. Medical Image Computing and Computer Assisted Intervention (MICCAI)},
publisher = {Springer},
}
The IJCARS paper describes the integration of tool modeling and provides quantitative results:
@article{DeepDRR2019,
author = {Unberath, Mathias and Zaech, Jan-Nico and Gao, Cong and Bier, Bastian and Goldmann, Florian and Lee, Sing Chun and Fotouhi, Javad and Taylor, Russell and Armand, Mehran and Navab, Nassir},
title = {{Enabling Machine Learning in X-ray-based Procedures via Realistic Simulation of Image Formation}},
year = {2019},
journal = {International journal of computer assisted radiology and surgery (IJCARS)},
publisher = {Springer},
}
For the original DeepDRR, released alongside our 2018 paper, please see the Version 0.1.
CUDA Cubic B-Spline Interpolation (CI) used in the projector:
https://github.com/DannyRuijters/CubicInterpolationCUDA
D. Ruijters, B. M. ter Haar Romeny, and P. Suetens. Efficient GPU-Based Texture Interpolation using Uniform B-Splines. Journal of Graphics Tools, vol. 13, no. 4, pp. 61-69, 2008.
The projector is a heavily modified and ported version of the implementation in CONRAD:
https://github.com/akmaier/CONRAD
A. Maier, H. G. Hofmann, M. Berger, P. Fischer, C. Schwemmer, H. Wu, K. Müller, J. Hornegger, J. H. Choi, C. Riess, A. Keil, and R. Fahrig. CONRAD—A software framework for cone-beam imaging in radiology. Medical Physics 40(11):111914-1-8. 2013.
Spectra are taken from MCGPU:
A. Badal, A. Badano, Accelerating Monte Carlo simulations of photon transport in a voxelized geometry using a massively parallel graphics processing unit. Med Phys. 2009 Nov;36(11): 4878–80.
The segmentation pipeline is based on the Vnet architecture:
https://github.com/mattmacy/vnet.pytorch
F. Milletari, N. Navab, S-A. Ahmadi. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. arXiv:160604797. 2016.
We gratefully acknowledge the support of the NVIDIA Corporation with the donation of the GPUs used for this research.