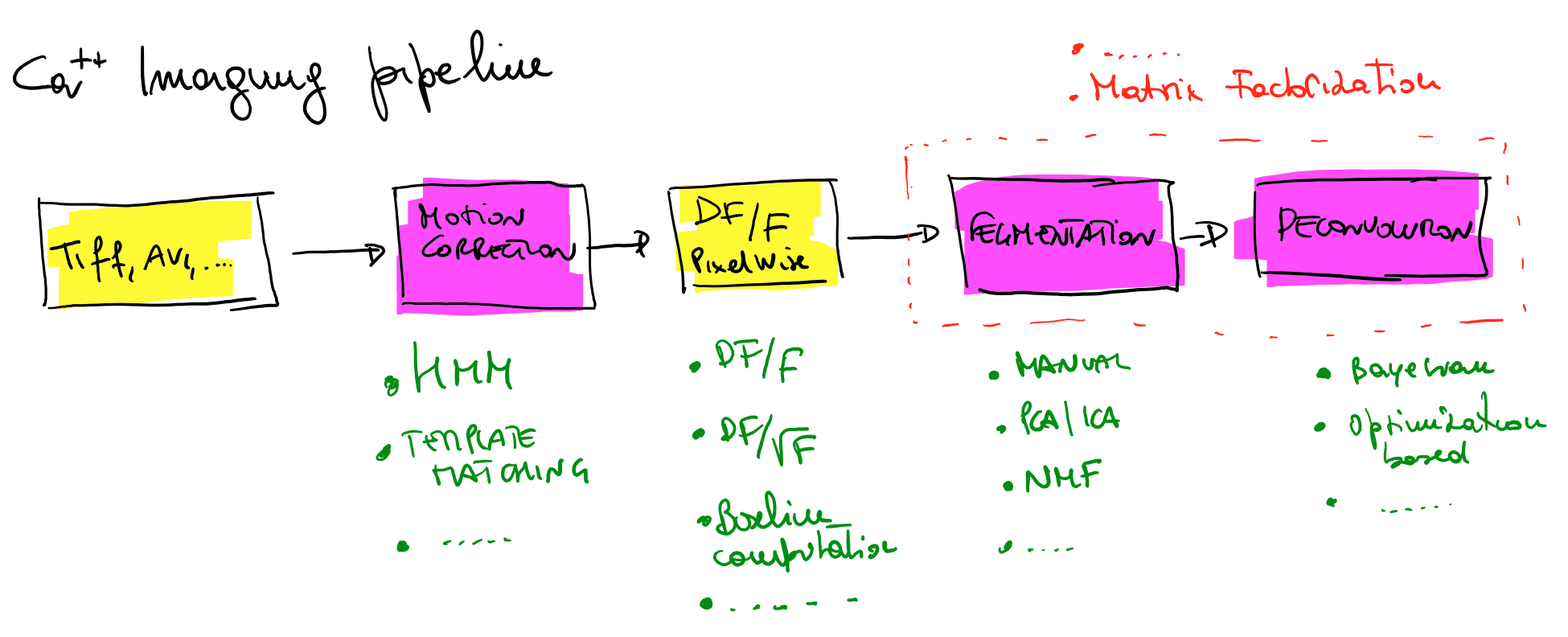
Blazing fast calcium imaging analysis toolbox
Recent advances in calcium imaging acquisition techniques are creating datasets of the order of Terabytes/week. Memory and computationally efficient algorithms are required to analyze in reasonable amount of time terabytes of data. This projects implements a set of essential methods required in the calcium imaging movies analysis pipeline. Fast and scalable algorithms are implemented for motion correction, movie manipulation and roi segmentation. It is assumed that movies are collected with the scanimage data acquisition software and stored in .tif format. Find below a schematic of the calcium imaging pipeline:
#%% initialization
import calblitz as cb
import time
import pylab as pl
import numpy as np
initTime = time.time()
filename = 'movies/demoMovie_PC.tif'
filename_hdf5 = filename[:-4]+'.hdf5'
#%% load and motion correct movie (see other Demo for more details)
print 'Loading data...'
frameRate = 15.62
start_time = 0
m = cb.load(filename, fr=frameRate, start_time=start_time)
print 'elapsed time:' + str(time.time()-initTime)
#%% automatic parameters motion correction
print 'Motion correction...'
max_shift_h = 10
max_shift_w = 10
m, shifts, xcorrs, template = m.motion_correct(max_shift_w=max_shift_w,
max_shift_h=max_shift_h,
num_frames_template=None,
template=None,
method='opencv')
max_h, max_w = np.max(shifts, axis=0)
min_h, min_w = np.min(shifts, axis=0)
m = m.crop(crop_top=max_h,
crop_bottom=-min_h+1,
crop_left=max_w,
crop_right=-min_w,
crop_begin=0,
crop_end=0)
print 'elapsed time:' + str(time.time()-initTime)
#%% play movie
print 'Playing movie, press q to stop...'
m.play(fr=50, gain=3.0, magnification=1)
#%% resize to increase SNR and convergence of segmentation algorithms
print 'Resizing data...'
resizeMovie = False
if resizeMovie:
fx = .5 # downsample a factor of two along x axis
fy = .5 # downsample a factor of two along y axis
fz = .2 # downsample a factor of 5 across time dimension
m = m.resize(fx=fx, fy=fy, fz=fz)
else:
fx, fy, fz = 1, 1, 1
print 'elapsed time:' + str(time.time()-initTime)
#%% debleach the signal by fitting a model on median intensity
print 'Debleaching data...'
m = m.debleach()
print 'elapsed time:' + str(time.time()-initTime)
#%% compute delta f over f (DF/F)
print 'Computing DF/F...'
m = m - np.min(m) + 1
m, mbl = m.computeDFF(secsWindow=10, quantilMin=50)
print 'elapsed time:' + str(time.time()-initTime)
#%% denoise and local correlation. this makes the movie look much better
print 'Denoising data...'
loc_corrs = m.local_correlations(eight_neighbours=True)
m = m.IPCA_denoise(components=100, batch=100000)
m = m * loc_corrs
print 'elapsed time:' + str(time.time()-initTime)
#%% compute spatial components via ICA PCA
print 'Computing PCA + ICA...'
spcomps = m.IPCA_stICA(componentsPCA=70, componentsICA=50,
mu=1, batch=1000000, algorithm='parallel',
whiten=True, ICAfun='logcosh', fun_args=None,
max_iter=2000, tol=1e-8, w_init=None, random_state=None)
print 'elapsed time:' + str(time.time()-initTime)
cb.matrixMontage(spcomps, cmap=pl.cm.gray) # visualize components
#%% extract ROIs from spatial components
print 'Extracting ROIs...'
_masks, _ = cb.extractROIsFromPCAICA(spcomps, numSTD=4.0,
gaussiansigmax=.1, gaussiansigmay=.2)
cb.matrixMontage(np.asarray(_masks), cmap=pl.cm.gray)
#%% extract single ROIs from each mask
minPixels, maxPixels = 20, 400
masks_tmp = []
for mask in _masks:
numPixels = np.sum(np.array(mask))
if (numPixels > minPixels and numPixels < maxPixels):
# print numPixels
masks_tmp.append(mask > 0)
masks_tmp = np.asarray(masks_tmp, dtype=np.float16)
all_masksForPlot_tmp = [kk*(ii+1)*1.0 for ii, kk in enumerate(masks_tmp)]
len(all_masksForPlot_tmp)
print 'elapsed time:' + str(time.time()-initTime)
#%% reshape dendrites if required (if the movie was resized)
if fx != 1 or fy != 1:
mdend = cb.movie(np.array(masks_tmp, dtype=np.float32), fr=1)
mdend = mdend.resize(fx=1/fx, fy=1/fy)
all_masks = mdend
else:
all_masks = masks_tmp
all_masksForPlot = [kk*(ii+1)*1.0 for ii, kk in enumerate(all_masks)]
#%% final visualization
print 'Visualization...'
mask_show = np.max(np.asarray(all_masksForPlot_tmp, dtype=np.float16), axis=0)
loc_corrs = m.local_correlations(eight_neighbours=True)
pl.imshow(loc_corrs, cmap=pl.cm.gray, vmin=0.5, vmax=1)
pl.imshow(mask_show > 0, alpha=.3, vmin=0)
cb.matrixMontage(np.asarray(all_masksForPlot_tmp), cmap=pl.cm.gray)
print 'Total elapsed time:' + str(time.time()-initTime)
LINUX
install anaconda python distribution, then in your terminal type
conda create --name calblitz python=2.7 python matplotlib scipy ipython h5py
source activate calblitz
conda install pip
conda install scikit-learn (or pip install scikit-learn)
conda install scikit-image
pip install pims
conda install -c menpo opencv3=3.1.0
pip install tifffile
conda install ipyparallel
conda install scipy
pip install tqdm
MAC OS X
install anaconda python distribution, then in your terminal type
conda create --name calblitz python=2.7 python matplotlib scipy ipython h5py
source activate calblitz
conda install pip
conda install scikit-learn (or pip install scikit-learn)
conda install scikit-image
pip install pims
pip install tifffile
conda install -c menpo opencv3=3.1.0
conda install ipyparallel
conda install scipy
pip install tqdm
WINDOWS
install anaconda python distribution, then in your terminal type
conda create --name calblitz python=2.7 python matplotlib scipy ipython h5py
activate calblitz
conda install pip
conda install scikit-learn (or pip install scikit-learn)
conda install scikit-image
pip install pims
pip install tifffile
conda install -c menpo opencv3=3.1.0
conda install ipyparallel
conda install scipy
pip install tqdm
if this does not work, you need to manually install opencv (pain in the neck)
For opencv windows installation check here
If you have problems installing opencv remember to match your architecture (32/64 bits) and to make sure that you have the required libraries installed
clone the git package
git clone https://github.com/agiovann/CalBlitz.git
or download the zipped version
cd CalBlitz/
type
python test_software.py
Add the CalBlitz folder to your Python path (or call the script from within the library). We suggest to use spyder to run the example code in DemoMotionCorrection.py or *DemoSegmentation.py. Each code cell is a unit that should be run and the result inspected. This package is supposed to be used interactively, like in MATLAB.
TODO
Andrea Giovannucci, Ben Deverett, Chad Giusti
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.
- Depending on terminal program used anaconda may not be in default path. In this case, add anaconda to bin to path:
export PATH=//anaconda/bin:$PATH
- Error: No packages found in current osx64 channels matching: pims install pip: conda install pip use pip to install pims: pip install pims if pims causes kernel crash then use
pip install pims --upgrade- If you get another compile time error installing pims, install the following
Microsoft C++ ompiler for Python
by typing
msiexec /i <path to downloaded MSI File> ALLUSERS=1
- If you get an "insecure string pickle" ValueError, install libav and mplayer. On Mac OS X, this can be easily done with brew:
brew install libav
brew install mplayer
```