Email: hoelzer.martin@gmail.com
2020-05-04: This repository is now integrated into the EBI repository of the VIRify pipeline and will not be further maintained in this spot.
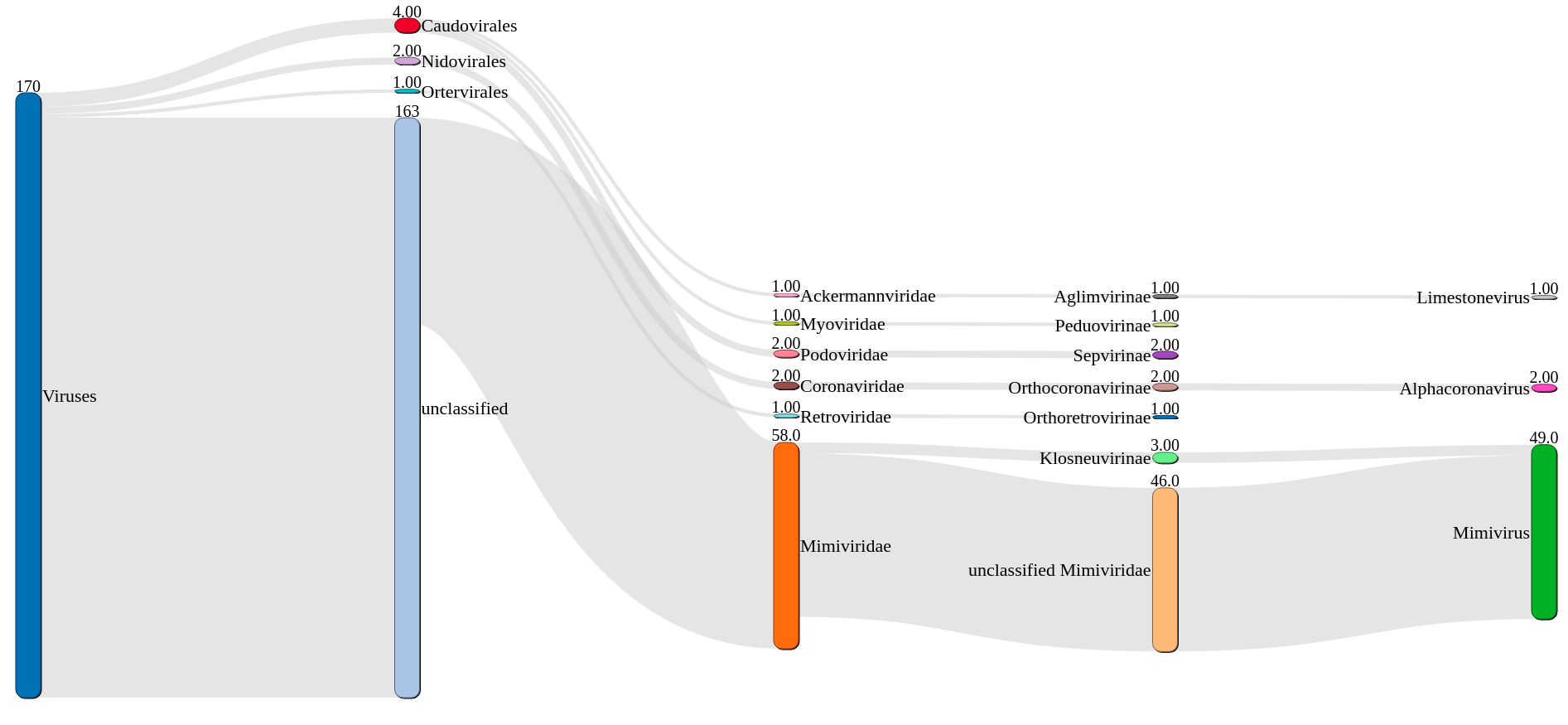
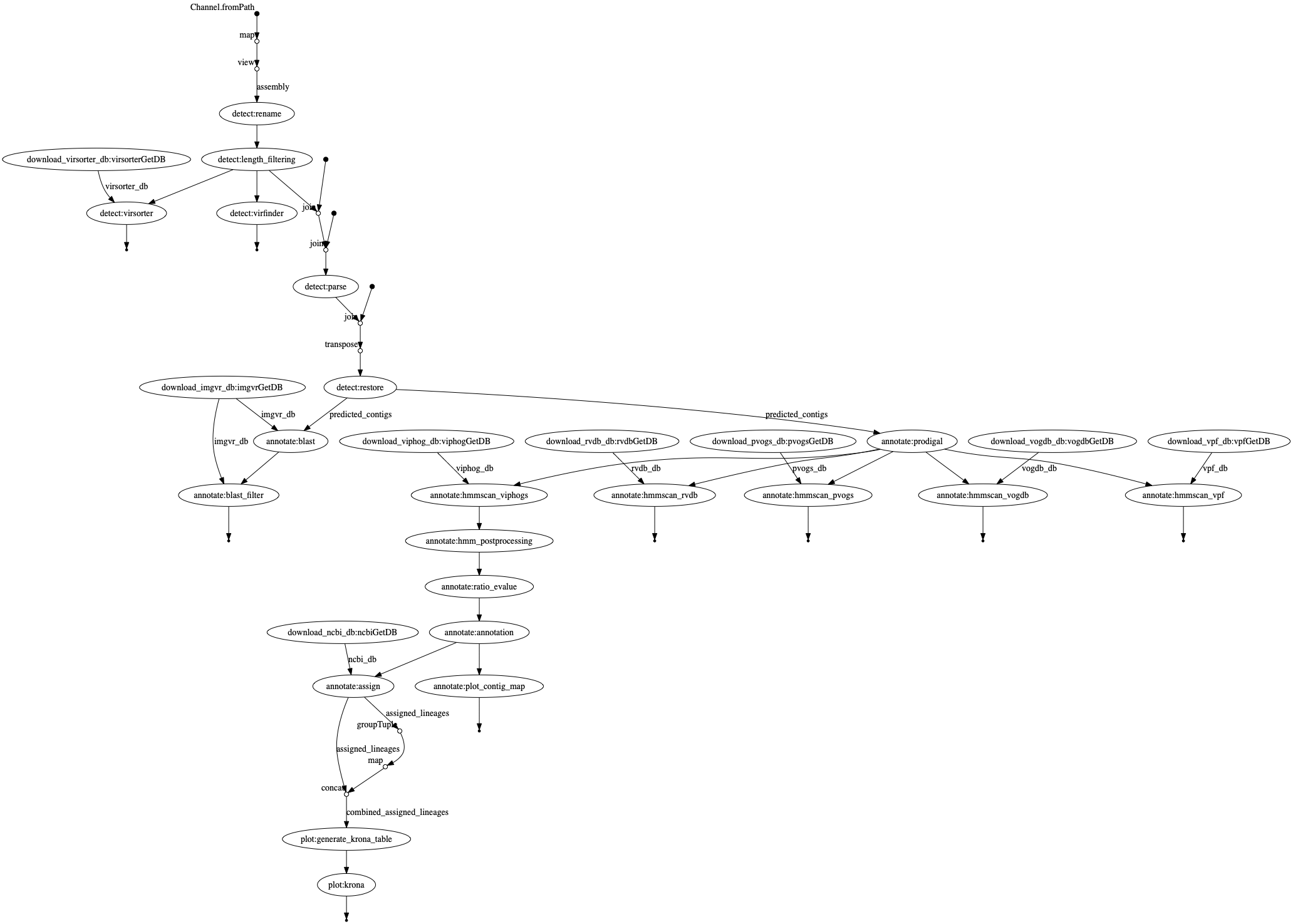
A nextflow implementation of the EBI VIRify pipeline for the detection of viruses from metagenomic assemblies. This implementation is heavily based on scripts and work by Guillermo Rangel-Pineros and the EBI Sequence Families Team.
This pipeline runs with the workflow manager Nextflow using Docker (Conda will be implemented soonish, hopefully). All other programs and databases are automatically downloaded by Nextflow. Attention, the workflow will download databases with a size of roughly 27 GB the first time it is executed.
curl -s https://get.nextflow.io | bashIf you dont have experience with bioinformatic tools and their installation just copy the commands into your terminal to set everything up:
sudo apt-get install -y docker-ce docker-ce-cli containerd.io
sudo usermod -a -G docker $USER- restart your computer
- see more instructions about Docker
Simply clone this repository or get or update the workflow via Nextflow:
nextflow pull hoelzer/virifyGet help:
nextflow run hoelzer/virify --helpRun annotation for a small assembly file (takes approximately 30min + time for database download; ~27 GB):
nextflow run hoelzer/virify --fasta '~/.nextflow/assets/hoelzer/virify/example_data/assembly.fasta'
Per default the workflow is run with Docker-support. When you execute the workflow on a HPC you can switch to
- SLURM (
-profile slurm) - LSF (
-profile lsf) and then you should also define the parameters - --workdir (here your work directories will be save)
- --databases (here your databases will be saved and the workflow checks if they are already available)
- --cachedir (here Docker/Singularity containers will be cached)
The -profile conda is not working at the moment. Sorry. Use Docker. Please.