Use job::job() to run chunks of R code in an RStudio job instead of the console. This frees your console while the job(s) go brrrrr in the background. By default, the result is returned to the global environment when the job completes.
Install:
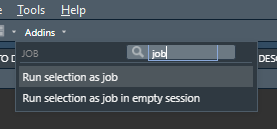
remotes::install_github("lindeloev/job")Two RStudio Addins are installed with job. Simply select some code code in your editor and click one of the Addins to run it as a job. The results are returned once the job completes.
- "Run selection as job" imports everything from your environment, so it feels like home. It will only return variables names that are not already in your environment upon submitting the job, so assign to novel names if you want them to return.
- "Run selection as job in empty session" imports nothing from your environment, so the code can run in clean isolation from the mess of a long-running session. All variables are returned, and overwritten if they already exist in your environment.
Write your script as usual. Then wrap parts of it using job::job({<your code>}) to run that chunk as a job:
job::job({
foo = 10
bar = rnorm(5)
})When the job completes, it silently saves foo and bar to your global environment.
Another use case: We all love brms but compilation and sampling takes time. Let's run it as a job!
library(brms)
data = mtcars[mtcars$hp > 100, ]
model = mpg ~ hp * wt
# Send long-running code to a job
job::job(brm_result = {
fit = brm(model, data)
fit = add_criterion(fit, "loo")
print(summary(fit)) # Show a summary in the job
the_test = hypothesis(fit, "hp > 0")
})
cat("I'm free now! Thank you.
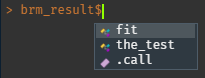
Sincerely, Console.")Now you can follow the progress in the jobs pane and your console is free. Because we named the code block brm_result, it will return the contents as an environment() called brm_result (or whatever you called it).brm_result behaves much like a list():
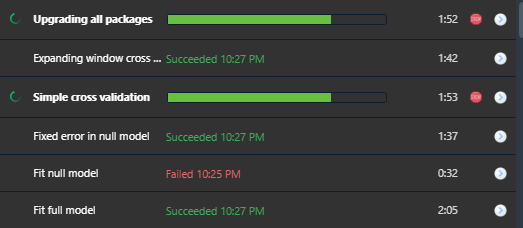
Often, the results of the long-running chunks are the most interesting. But they easily get buried among the other outputs in the console and are eventually lost due to the line limit. RStudio's jobs history can be used as a nice record. Make sure to print informative results within the job and give your jobs an appropriate title, e.g., (job::job({<code here>}, title = "Unit test: first attempt at feature X").
RStudio jobs spin up a new session, i.e., a new environment. By default, job::job() will make this environment identical to your current one. But you can fine control this:
-
import: the default "auto" setting imports all objects that are referenced by the code into the job. Control this usingjob::job({}, import = c(model, data)). You can also import everything (import = "all") or nothing (import = NULL). -
packages: by default, all attached packages are attached in the job. Control this usingjob::job({}, packages = c("brms"))or setpackages = NULLto load nothing. Ifbrmsis not loaded in your current session, addinglibrary("brms")to the job code may be more readable. -
options: by default, all options are overwritten/inserted to the job. Control this using, e.g.,job::job({}, opts = list(mc.cores = 2)or setopts = NULLto use default options. If you want to set job-specific options, addingoptions(mc.cores = 2)to the job code may be more readable. -
export: in the example above, we assigned the job environment tobrm_resultupon completion. Naturally, you can choose any name, e.g.,job::job(fancy_name = {a = 5}). To return nothing, use an unnamed code chunk (insert results toglobalenv()and remove everything before return: (job::job({a = 5; rm(list=ls())}). Returning nothing is useful when-
your main result is a text output or a file on the disk, or
-
when the return is a very large object. The underlying
rstudioapi::jobRunScript()is slow in the back-transfer so it's usually faster tosaveRDS(obj, filename)them in the job andreadRDS(filename)into your current session.
-
- Model training, cross validation, or hyperparameter tuning: train multiple models simultaneously, each in their own job. If one fails, the others continue.
- Heavy I/O tasks, like processing large files. Save the results to disk and return nothing.
- Run unit tests and other code in an empty environment. By default,
devtools::test()runs in the current environment, including manually defined variables (e.g., from the last test-run) and attached packages. Calljob::job({devtools::test()}, import = NULL, packages = NULL, opts = NULL)to run the test in complete isolation. - Upgrading packages
job::job() is aimed at easing interactive development within RStudio. For larger problems, production code, and solutions that work outside of RStudio, check out:
-
The
futurepackage's%<-operator combined withplan(multisession). -
The
callrpackage is a general tool to run code in new R sessions.