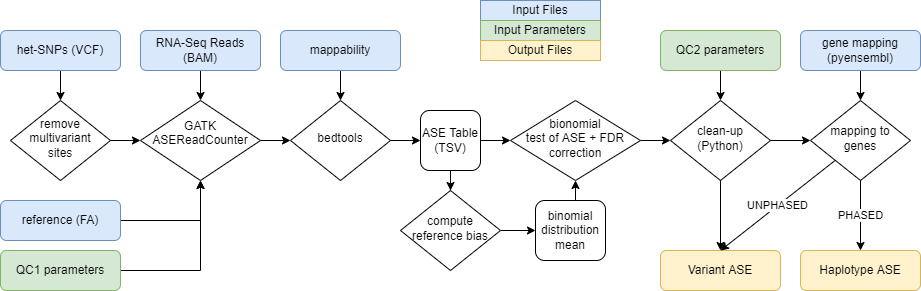
This is a ICGC-ARGO pipeline for analysis of allele specific expression (ASE) based on RNA-seq data.
The pipeline processes a reads file (BAM/SAM) and its accompanying variant call files (VCF) together with their matching reference (FA) file and produces, for each SNP, the allele-specific expression ratio and the probability that true ASE is occuring.Using a (GTF) file the positions are also mapped to genes.
If the data are additionally phased, the haplotype-specific expression for each gene is computed.
The whole pipeline operates as illustrated:
The pipeline can be modified using the following quality control parameters:
- QC paramters (applied before ASE read counter)
- read depth (
16) - read mapping quality (
20) - read calling quality (
10) - read mappability (
0.05)
- read depth (
For a file sample_name.bam we obtain the following outputs:
sample_name.tsv: tab separated document detailing the results of the ASE analysis with the following result columns:ase_ratio: the RAF adjusted for mean bias towards referenceref_bias: the ration of reference counts vs total read counts for the particular base pairAEI_pval: the resulting p-value of binomial statistical testAEI_padj: the p-value corrected using Benjamini/Hochberg false discovery rate correction. The AE is present ifp < 0.5.gene_id: if a genome file is provided, maps to an Ensembl gene idgene_feature: if a genome file is provided, maps to an Ensembl gene feature (exon/intron)
sample_name.gene.log: a log file of the ASE calculation and filteringsample_name.vaf.png: a histogram ofase_ratiooccurences. In a healthy sample the values should be around 0.5.sample_name.hap.tsv: if a genome file is provided and the data are phased, the results of ASE mapped to genes, the result colums are:positions: how many positions are covered by a geneHSE_ratio: ratio of the first haplotype vs. totalHEI_pval: the resulting p-value of binomial statistical test for the geneHEI_padj: the FDR B/H p-value correction
sample_name.hap.log: a log file of the haplotype specific expression calculation
The following human genome files have been tested with the pipeline:
https://object.cancercollaboratory.org:9080/swift/v1/genomics-public-data/rna-seq-references/GRCh38_Verily_v1.genome/GRCh38_Verily_v1.genome.fa.gz
https://bismap.hoffmanlab.org/raw/hg38/k50.umap.bedgraph.gz
https://object.cancercollaboratory.org:9080/swift/v1/genomics-public-data/rna-seq-references/GRCh38_Verily_v1.annotation/gencode.v40.chr_patch_hapl_scaff.annotation.gtf
Email questions, feature requests and bug reports to Adam Streck, adam.streck@mdc-berlin.de.
icgc-argo-workflows/allele-sepecific-expression is available under the MIT License.
Tools and best practices for data processing in allelic expression analysis, Castel et al.