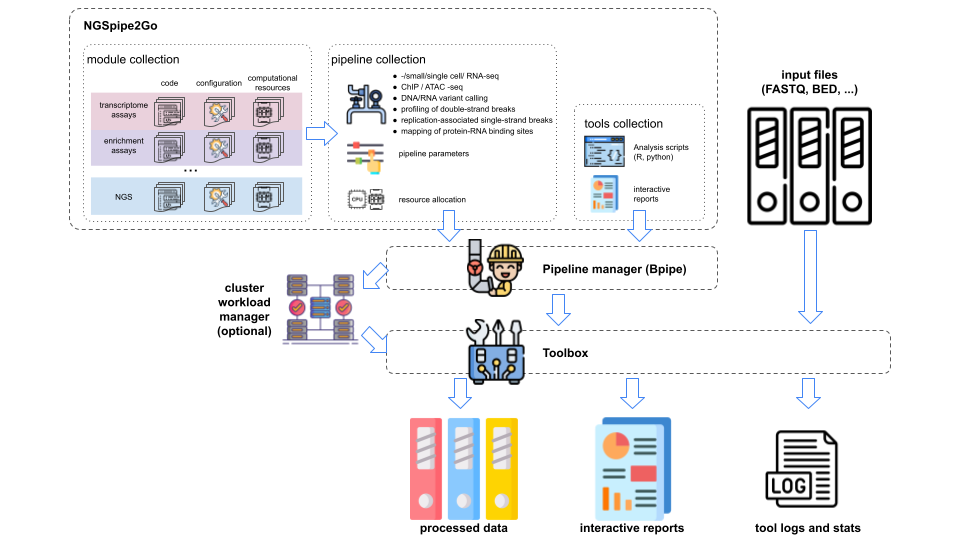
NGSpipe2go is a software framework to facilitate the development, customization and deployment of NGS analysis pipelines developed and utilised at the Bioinformatics Core of IMB in Mainz (https://www.imb.de/). It uses Bpipe as a workflow manager, provides access to commonly used NGS tools and generate comprehensive reports based on R Markdown.
NGS projects should be run in a consistent and reproducible way, hence NGSpipe2go asks you to copy all tools into the project folder, which will ensure that you always use the same program versions at a later time point. This can be done either from a local NGSpipe2go copy or by using the most recent version from the Gitlab repository
git clone https://gitlab.rlp.net/imbforge/NGSpipe2go <project_dir>/NGSpipe2go
Select a pipeline to run and make symlinks in the main project dir, e.g. for RNA-seq projects
ln -s NGSpipe2go/pipelines/RNAseq/* .
or for ChIP-seq projects
ln -s NGSpipe2go/pipelines/ChIPseq/* .
Adjust the project-specific information in the pipeline dependent files (see pipeline specific README files for detailed information):
- essential.vars.groovy specifies the main project variables like project dir and reference genome
- xxx.header additional software parameters can be customised in the vars-file accompanying each bpipe module.
- xxx.pipeline.groovy describes the steps of the selected pipeline and the location of the respective modules
- targets.txt and contrasts.txt contain the sample names and the differential group comparisons
Optionally adjust some general pipeline settings defined in the NGSpipe2go config folder:
- bpipe.config.groovy: define workload manager resources (default workload manager is "slurm", if not needed set executor="local")
- preambles.groovy: define module preambles if needed (or stay with default preambles)
- tools.groovy: define default versions and running environments for all installed pipeline tools, modify accordingly if new tools or tool versions are installed on your system. If you want to use a different tool version for a certain project you can overwrite the default value in the pipeline-specific file NGSpipe2go/pipelines//tools.groovy. Currently, we are using lmod, conda and/or singularity containers as running environments. Conda itself is loaded as lmod module if conda tools are specified. If you want to use conda but haven't installed it as lmod module make otherwise sure that it is available in the path.
Copy the input FastQ files in the <project_dir>/rawdata folder.
Load the bpipe module customised for the Slurm job manager (we recommend to use GNU Screen for persistence), e.g.
screen
module load bpipe/0.9.9.8.slurm
Start running the pipeline of choice, e.g.
bpipe run rnaseq.pipeline.groovy rawdata/*.fastq.gz
or
bpipe run chipseq.pipeline.groovy rawdata/*.fastq.gz
The results of the pipeline modules will be saved in the ./results folder. The final report Rmd-file is stored in the ./reports folder and can be edited or customised using a text editor before converting into a HTML report using knitr
R usage:
rmarkdown::render("DEreport.Rmd")
or
rmarkdown::render("ChIPreport.Rmd")
When deleting intermediate files and re-running the pipeline, attribute caching of NFS servers might lead to incorrectly seen time stamps of files and, thus, downstream files not being updated. This can be circumvented by either remove all downstream files before re-running or forcing all modules to be run on the same node as the bpipe instance itself is running. The latter can be achived by adding
custom_submit_options="--nodelist=NODENAME"
to the general options specified in the config file bpipe.config.groovy.