Our goal is to develop interpretive standards for AMR genotypes, akin to the interpretive standards developed by EUCAST and CLSI for antimicrobial susceptibility phenotyping.
An overview of the concept, with example data structures and code, is available below.
We are partnering with ESGEM, the ESCMID Study Group on Epidemiological Markers, to form an ESGEM-AMR Working Group to curate organism-specific rule sets. Membership of the working group is open to anyone with relevant expertise, you do not have to be an ESGEM member.
If you would like to get involved in the ESGEM-AMR Working Group, please go here.
Organism-specific interpretation of antimicrobial susceptibility testing (AST) data is standard in clinical microbiology, with rules regularly reviewed by expert committees of CLSI and EUCAST. EUCAST also maintains lists of expert rules for some species, including expected (intrinsic) resistance and expected susceptibility phenotypes, to guide clinical labs in deciding which drugs to test and whether/how to report them.
We propose there is a similar need for systematic rules for the organism-specific interpretation of antimicrobial resistance (AMR) genotypes derived from pathogen whole genome sequence (WGS) data.
Current solutions focus on bespoke solutions for specific organisms (e.g. our Kleborate tool for Klebsiella pneumoniae; Pathogenwatch AMR libraries for Salmonella Typhi, Neisseria gonorrhoeae and others; Resfinder 4.0 for E. coli and others; Mykrobe and TBProfiler tools for Mycobacterium tuberculosis), but this complicates bioinformatics analyses and promotes fragmentation rather than consolidation of expertise. AbritAMR offers a potential solution for multiple organisms, but organism-specific interpretation rules are hard-coded in Python and separated from supporting evidence, making the logic difficult for others to curate and update.
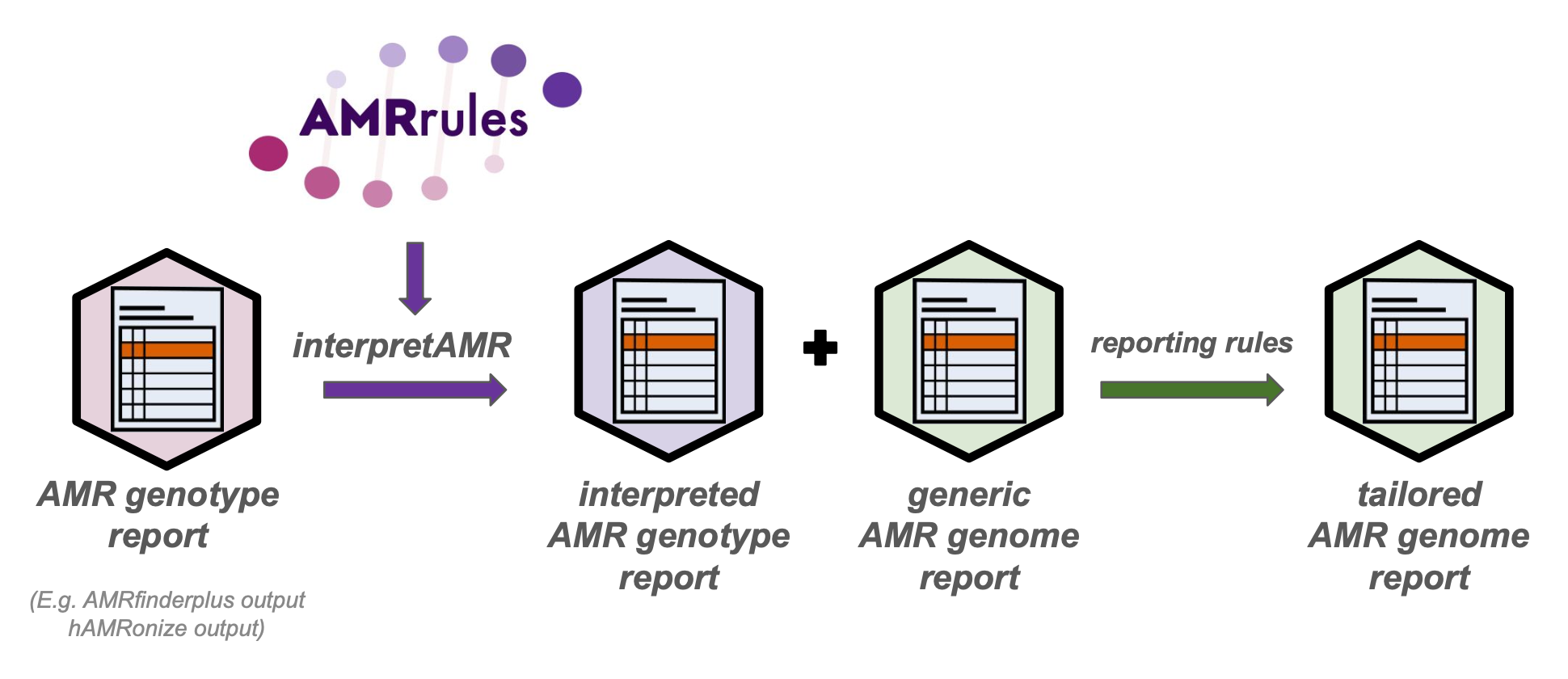
This repository outlines a proposal for a simple data structure to store organism-specific rules for the interpretation of AMR genotype data, that could be used to enrich the outputs of standard AMR genotyping tools (such as AMRfinderplus and other tools, with or without hAMRonization) and generate informative genome reports that capture expert knowledge about how core genes contribute to antimicrobial susceptibility.
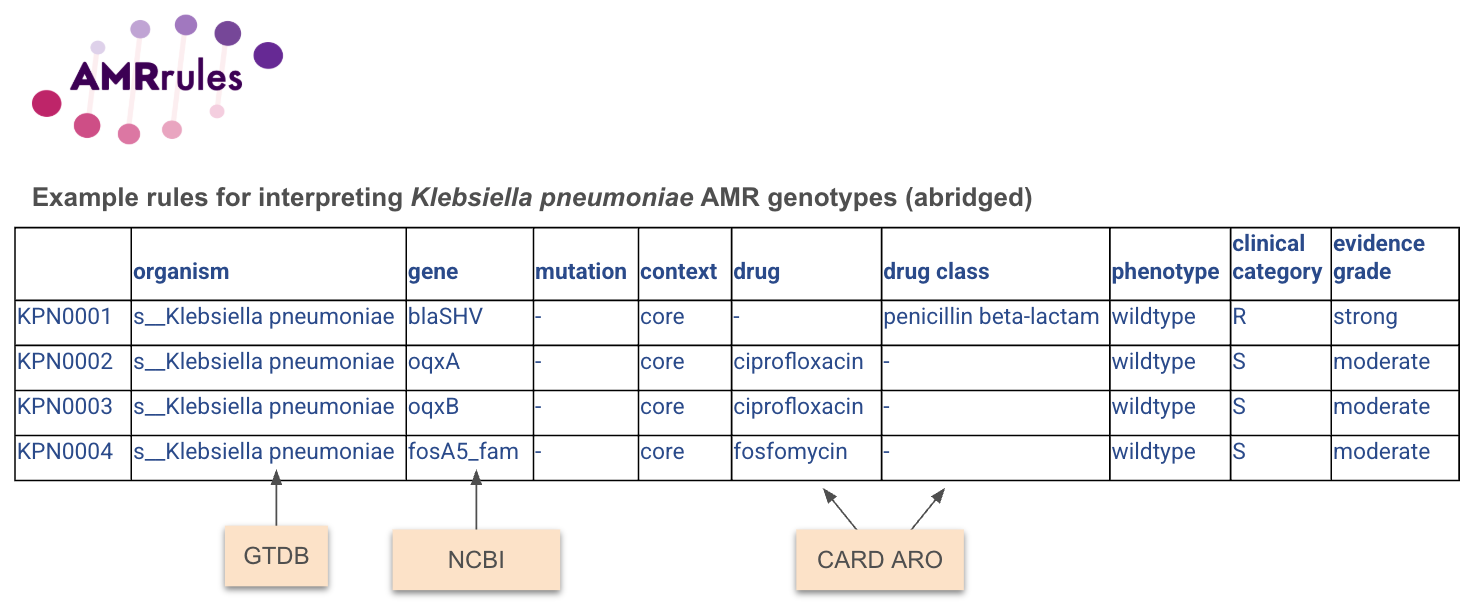
Example rules are given for Klebsiella pneumoniae, and example code is given to annotate NCBI AMRfinderplus results for Klebsiella pneumoniae using these rules.
The rule specification (still a work in progress) is available in this Google sheet (v0.2, guidance on tab 2).
We have partnered with ESGEM, the ESCMID Study Group on Epidemiological Markers, to form an ESGEM-AMR Working Group to curate organism-specific rule sets.
We are also partnering with EUCAST to ensure alignment of the AMRrules approach with the EUCAST Subcommittee on WGS and Phenotypic AST, including their first report (2017) and ongoing updates.
Example file: organism_specific_rules.tsv
Full specification (work in progress): AMR rules specification. Note this includes some additional fields, and guidance on tab 2.
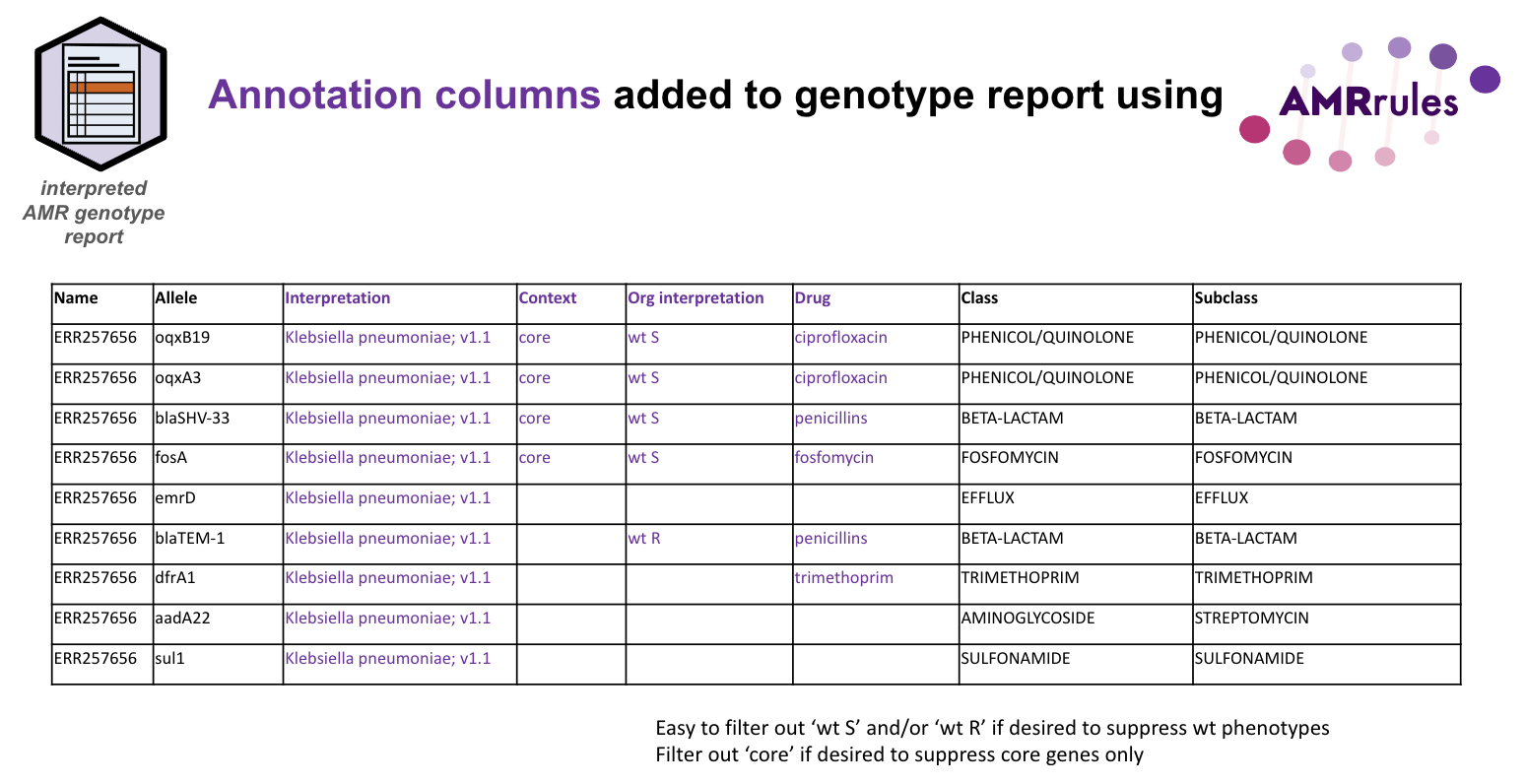
Example file: annotated_gene_report.tsv
The example annotated gene report can be generated by running AMRfinderplus, and then annotating the output with the above organism-specific rules for Klebsiella pneumoniae, using parse_amrfinder.py
amrfinder -n example_data_kleb/ERR257656.fasta --plus
--print_node
--name ERR257656
--organism Klebsiella_pneumoniae
> ERR257656_amrFinderPlusOutput.tsv
./parse_amrfinder.py --reports example_data_kleb/*_amrFinderPlusOutput.tsv
--species 'Klebsiella pneumoniae'
--organism_rules organism_specific_rules.tsv
--drug_dictionary example_dict_kleb/Kleb_local_dict.tsv
--output example_annotatedAMRreport.tsv
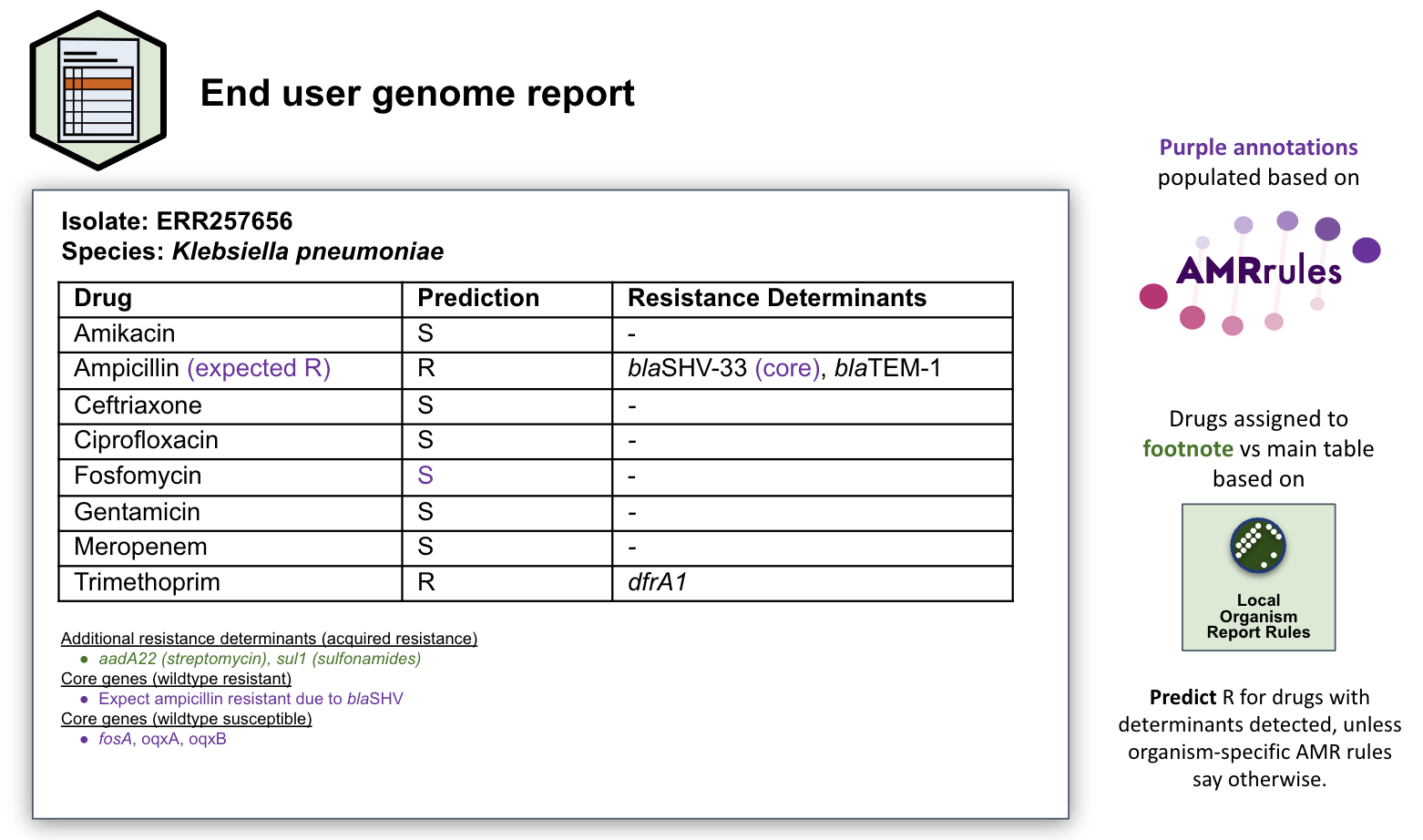
Example file (PDF): genome_report.pdf
Example file (RTF): genome_report.rtf
This concept was workshopped by members of the Holt lab at London School of Hygiene and Tropical Medicine and further developed in collaboration with Jane Hawkey at Monash University.