mapgen is an R package that performs gene mapping based on
functionally-informed genetic fine-mapping.
You can install the development version of mapgen from
GitHub with:
# install.packages("remotes")
remotes::install_github("kevinlkx/mapgen")- Please install
susieRpackage, if you want to run finemapping with GWAS summary statistics using SuSiE. - Please install TORUS software package, if you want to run enrichment analysis using TORUS.
After installing, check that it loads properly:
library(mapgen)Example workflow from our heart single-cell study:
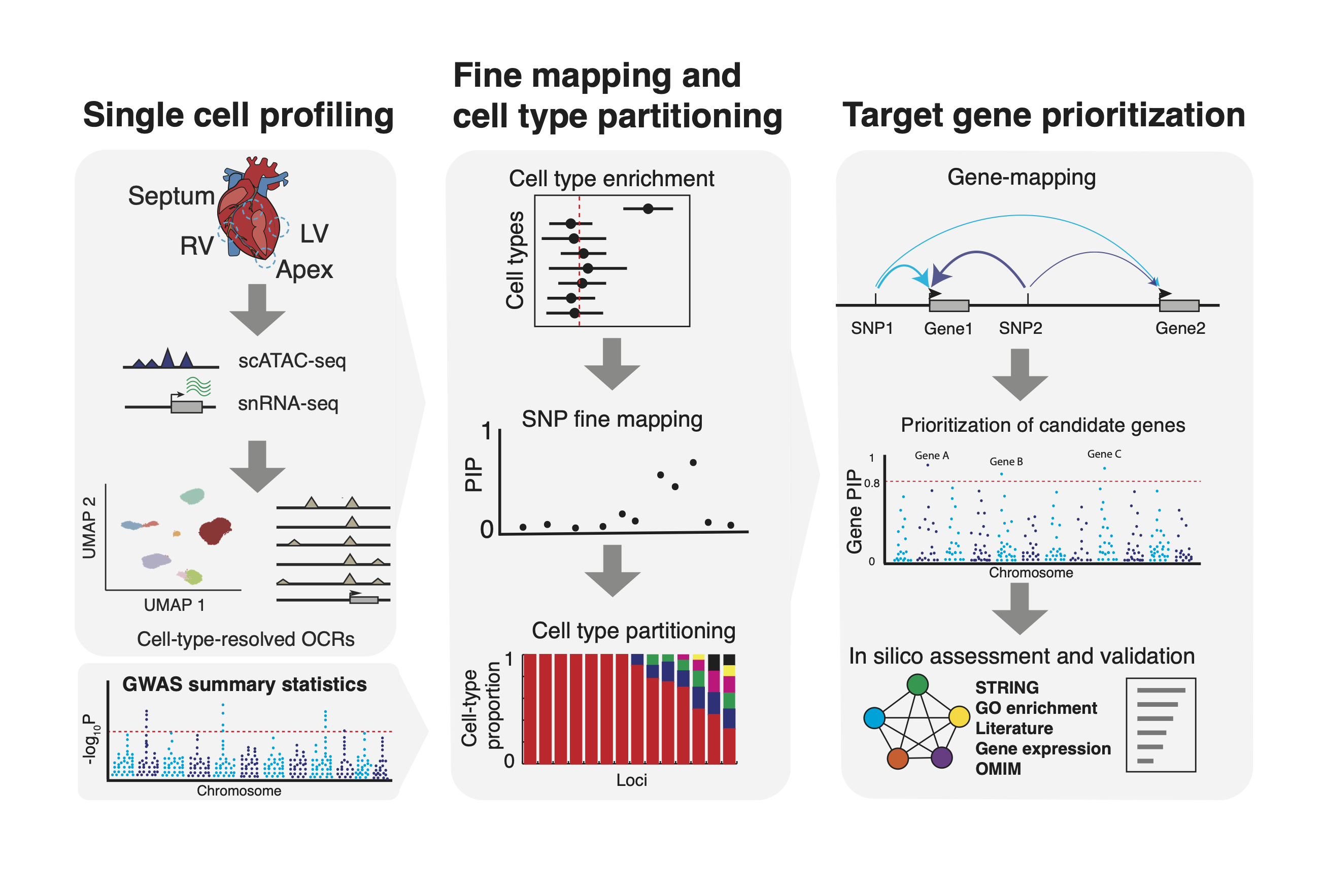
We developed an integrated procedure that combines single-cell genomics with novel computational approaches to study genetics of complex traits.
Main steps:
- Obtain cell-type-resolved open chromatin regions (OCRs) using scATAC-seq and snRNA-seq.
- Assess the enrichment of genetic signals of a trait of interest in OCRs across all the cell types.
- Perform Bayesian statistical fine mapping on trait-associated loci, using a informative prior that favors likely functional variants located in OCRs of enriched cell types.
- Assign the likely cell type(s) through which the causal variants act in most loci using fine-mapped SNPs and its associated cell type information.
- Use our novel gene mapping procedure to infer causal genes at each locus.
Please follow the tutorials to learn how to use the package.
Single-cell genomics improves the discovery of risk variants and genes of cardiac traits. Alan Selewa*, Kaixuan Luo*, Michael Wasney, Linsin Smith, Chenwei Tang, Heather Eckart, Ivan Moskowitz, Anindita Basu, Xin He, Sebastian Pott. medRxiv 2022.02.02.22270312; doi: https://doi.org/10.1101/2022.02.02.22270312