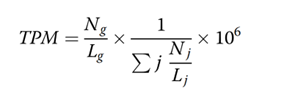The metagenomic clean reads were assembled to contigs using metaSPAdes with parameter "-meta –threads 40 -k 21,33,55,77,99,127", MetaProdigal was employed for gene prediction of the assembled contigs.
We used PlasForest[12] a homology-based random-forest classifier and PlasClass (parameter: score ≥ 0.99 and minimal contig length ≥ 500bp), a kmer-based logistic regression classifier to identify plasmid sequences in assembled contigs. The plasmid contigs were aligned to NCBI Refseq [14] plasmid database to identify the taxonomy origin using BLASTN (version 2.10.1) [15].
All clean reads were aligned to assembled contigs and predicted ORF with Bowtie2 (parameter: --end-to-end --sensitive -I 200 -X 400), ORFs were quantified with transcripts per million (TPM), TPM is calculated as:
where Ng is the read count, the reads number mapped to the g gene, and Lg is the gene length. The index j stands for the set of all predicted gene in sample, and g is an index indicating a particular gene.
CoverM was used for contigs abundance quantification.
| #software | versions | link |
|---|---|---|
| FastQC | 0.11.9 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic | 0.39 | http://www.usadellab.org/cms/?page=trimmomatic |
| BMTagger | 3.102 | ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger/ |
| MetaSPADes | 3.15.3 | https://github.com/ablab/spades |
| MetaProdigal | 2.6.3 | https://github.com/hyattpd/Prodigal |
| bowtie2 | 2.4.4 | https://github.com/BenLangmead/bowtie2 |
| coverm | 0.6.1 | https://github.com/wwood/CoverM |
| Kallisto | 0.46.2 | https://github.com/pachterlab/kallisto |
| mmseqs2 | r13 | https://github.com/soedinglab/MMseqs2 |
| PlasForest | 1.3 | https://github.com/leaemiliepradier/PlasForest |
| seqtk | 0.1 | https://github.com/lh3/seqtk |
| tabtk | 0.1 | https://github.com/lh3/tabtk |
| csv2tsv | 2.2.0 | https://github.com/eBay/tsv-utils |
| R | 4.2.1 | https://www.r-project.org/ |
- Andrews, S., FASTQC. A quality control tool for high throughput sequence data. 2010.
- Bolger, A.M., M. Lohse, and B. Usadel, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England), 2014. 30(15): p. 2114-2120.
- Rotmistrovsky, K. and R. Agarwala, BMTagger: Best Match Tagger for removing human reads from metagenomics datasets. Unpublished, 2011.
- Nurk, S., et al., metaSPAdes: a new versatile metagenomic assembler. Genome research, 2017. 27(5): p. 824-834.
- Bray, N.L., et al., Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology, 2016. 34(5): p. 525-527.
- Alcock, B.P., et al., CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research, 2020. 48(D1): p. D517-D525.
- Steinegger, M. and J. Söding, MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nature Biotechnology, 2017. 35(11): p. 1026-1028.
- Pradier, L., et al., PlasForest: a homology-based random forest classifier for plasmid detection in genomic datasets. BMC Bioinformatics, 2021. 22(1): p. 349.
- Pellow, D., I. Mizrahi, and R. Shamir, PlasClass improves plasmid sequence classification. PLOS Computational Biology, 2020. 16(4): p. e1007781.
- Kitts, P.A., et al., Assembly: a resource for assembled genomes at NCBI. Nucleic acids research, 2016. 44(D1): p. D73-D80.
- Dixon, P., VEGAN, a package of R functions for community ecology. Journal of Vegetation Science, 2003. 14(6): p. 927-930.