The AI-powered MD is a generalizable solution to efficiently simulate various proteins with ab initio accuracy by machine learning force field. This project consists of our studies on Datasets, Modeling, Simulation evaluation and analysis, which are demonstrated below and in different branches. See The Homepage of AI2BMD and find the preprint version article AI2BMD: efficient characterization of protein dynamics with ab initio accuracy for more details.
Hiring: We are hiring research interns, engineering interns and full time employees on MD simulation, quantum chemistry, AIDD, geometry deep learning (GDL), molecular graph neural network, system design and CUDA acceleration. Please send your resume to watong@microsoft.com .
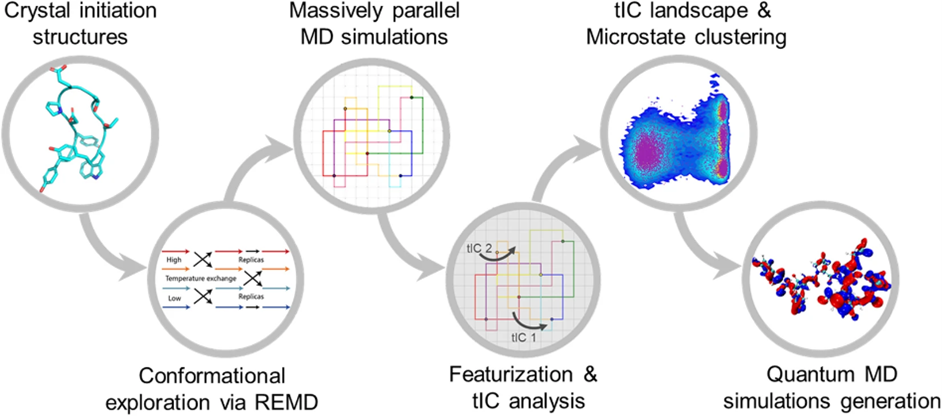
The whole comformation MD dataset for proteins calculated at Density Functional Theory (DFT) level. AIMD-Chig consists of 2M conformations of the 166-atom Chignolin and the corresponding potential energy and atomic forces calculated at M06-2X/6-31g* level.
-
Read the article AIMD-Chig: Exploring the conformational space of a 166-atom protein Chignolin with ab initio molecular dynamics.
-
Find the story The first whole conformational molecular dynamics dataset for proteins at ab initio accuracy and the novel computational technologies behind it.
-
Get the dataset AIMD-Chig.
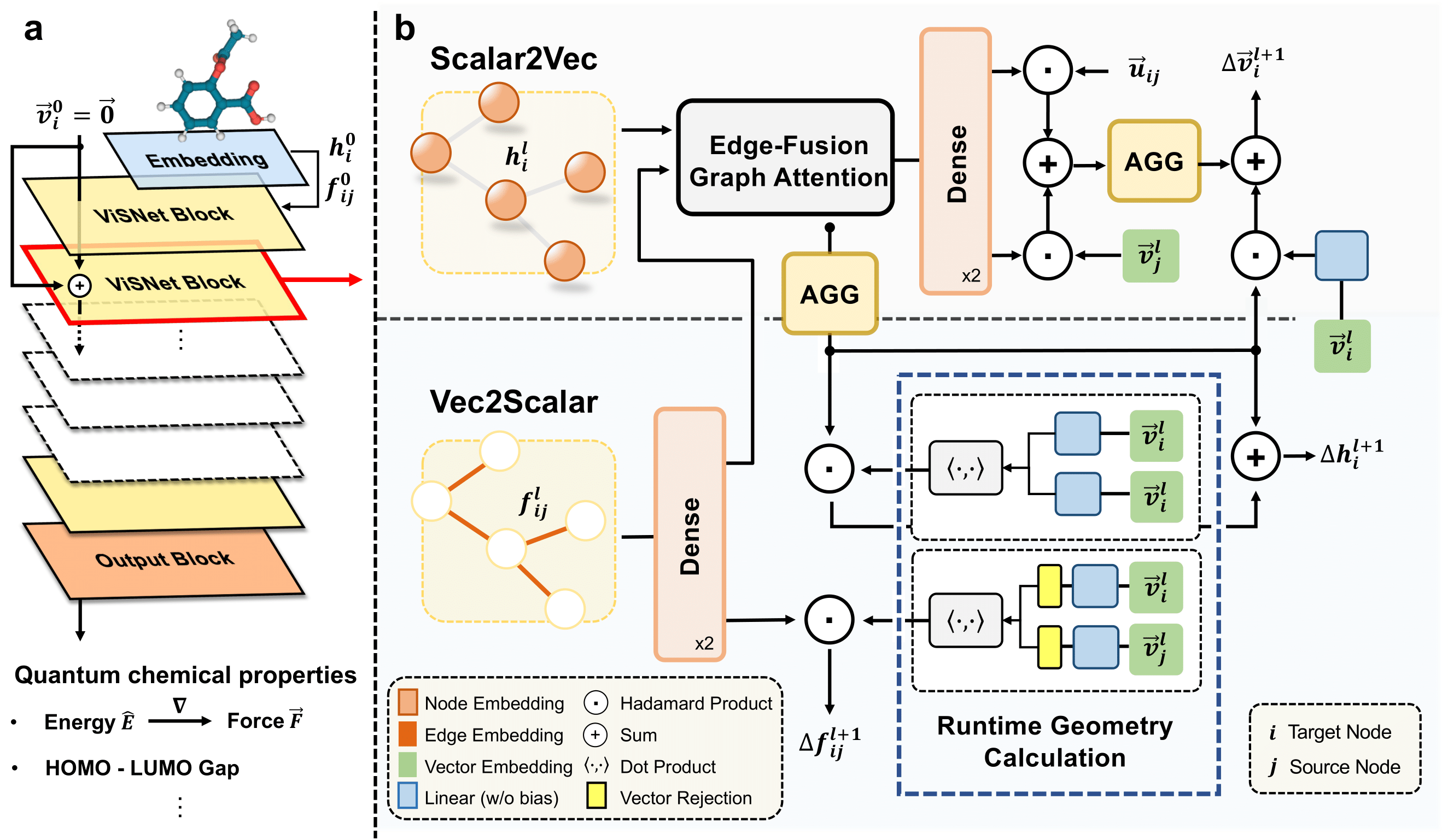
ViSNet (shorted for “Vector-Scalar interactive graph neural Network”) is an equivariant geometry-enhanced graph neural for molecules that significantly alleviate the dilemma between computational costs and sufficient utilization of geometric information. ViSNet has won the Championship in The First Global AI Drug Development Competition and one of the winners in OGB-LSC @ NeurIPS 2022 PCQM4Mv2 Track!
-
Please check out the branch ViSNet for the source code, instruction on model training and more techniqucal details.
-
Read the preprint version article ViSNet: an equivariant geometry-enhanced graph neural network with vector-scalar interactive message passing for molecules.
Machine learning force fields (MLFFs) have gained popularity in recent years as they provide a cost-effective alternative to ab initio molecular dynamics (MD) simulations. Despite a small error on the test set, MLFFs inherently suffer from generalization and robustness issues during MD simulations. To alleviate these issues, we propose global force metrics and fine-grained metrics from element and conformation aspects to systematically measure MLFFs for every atom and every conformation of molecules. Furthermore, the performance of MLFFs and the stability of MD simulations can be further improved guided by the proposed force metrics for model training, specifically training MLFF models with these force metrics as loss functions, fine-tuning by reweighting samples in the original dataset, and continued training by recruiting additional unexplored data.
- Read the Cover Story article Improving machine learning force fields for molecular dynamics simulations with fine-grained force metrics .
Markov state models (MSMs) play a key role in studying protein conformational dynamics. A sliding count window with a fixed lag time is widely used to sample sub-trajectories for transition counting and MSM construction. However, sub-trajectories sampled with a fixed lag time may not perform well under different selections of lag time, which requires strong prior practice and leads to less robust estimation. To alleviate it, we propose a novel stochastic method from a Poisson process to generate perturbative lag time for sub-trajectory sampling and utilize it to construct a Markov chain. Comprehensive evaluations on the double-well system, WW domain, BPTI, and RBD–ACE2 complex of SARS-CoV-2 reveal that our algorithm significantly increases the robustness and power of a constructed MSM without disturbing the Markovian properties. Furthermore, the superiority of our algorithm is amplified for slow dynamic modes in complex biological processes.
-
Read the Cover Story article Stochastic Lag Time Parameterization for Markov State Models of Protein Dynamics.
-
Find an application case in studying the Spike-ACE2 complex structure for the highly infectious mechanism of Omicron: Structural insights into the SARS-CoV-2 Omicron RBD-ACE2 interaction.
Please contact Dr. Tong Wang (watong@microsoft.com) if you have interests in our study.
This project is licensed under the terms of the MIT license.