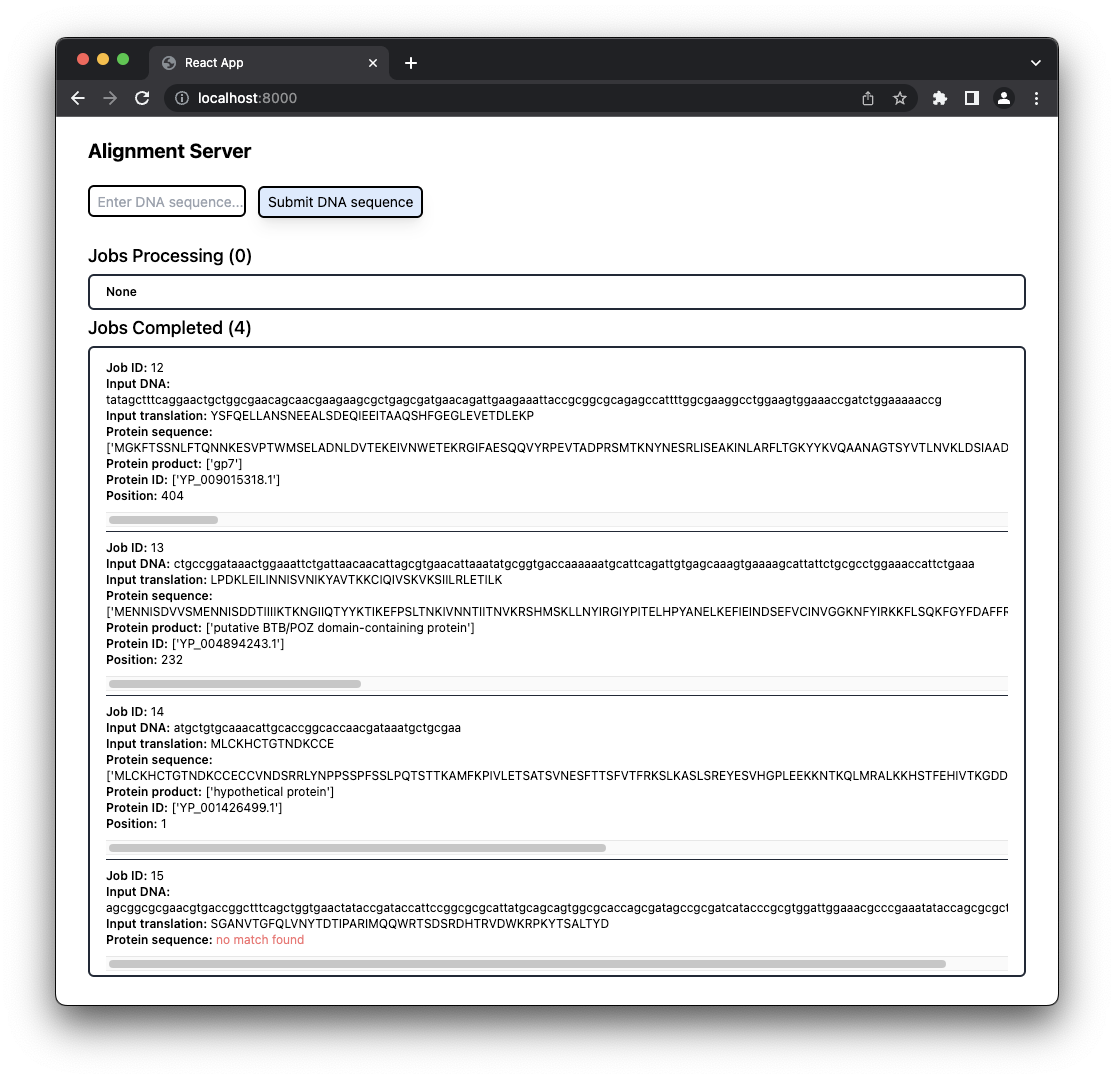
This project was a coding challenge to integrate Django and React and then deploy the site. The site translates input DNA to a peptide and searches genomes for a parent protein.
-
The client takes a DNA sequence as an input.
-
Each submission is sent to the backend, translated to protein, and the alignment is processed using a Python thread.
-
A set of ten genomes is searched. The genomes are in GenBank (.gb) format and included with the project.
-
Multiple submissions can be made on the client before the first submission has completed. The jobs in progress are displayed in the browser.
-
As each job completes it is shown in the browser with the results, including the name of the protein where the peptide was found, and the residue number where the match starts.
-
If the browser is closed the results will be automatically reloaded when the page is reopened.
-
Previous searches and results are visible.
-
The search is done by iterating through proteins and returning the first protein having a substring that matches the peptide sequence. This approach was chosen over an BioPython alignment because it yields results meeting the project specification, and could be implemented with the Python find() function which is simple and efficient.
-
While one or more jobs are in the queue the client polls the server. Polling is off when no jobs are in the queue. The use of polling is less efficient than using a websocket, and would likely not be suitable for a server that gets significant traffic. However, for the small scale of the demo project it seemed like a good approach.
-
Currently the app does limited input validation. This would need to be more robust in production.
-
The app assumes that one user or a unified team of users will submit jobs. For multiple users/teams it would be best to compartmentalize the different sets of results.
-
I chose PythonAnywhere to deploy because it's fast to set up, and it supports SQLite which is what I used for the project.