Welcome to the NEAT project, the NExt-generation sequencing Analysis Toolkit, version 3.2. Neat has now been updated with Python 3, and is moving toward PEP8 standards. There is still lots of work to be done. See the ChangeLog for notes.
Stay tuned over the coming weeks for exciting updates to NEAT, and learn how to contribute yourself. If you'd like to use some of our code, no problem! Just review the license, first.
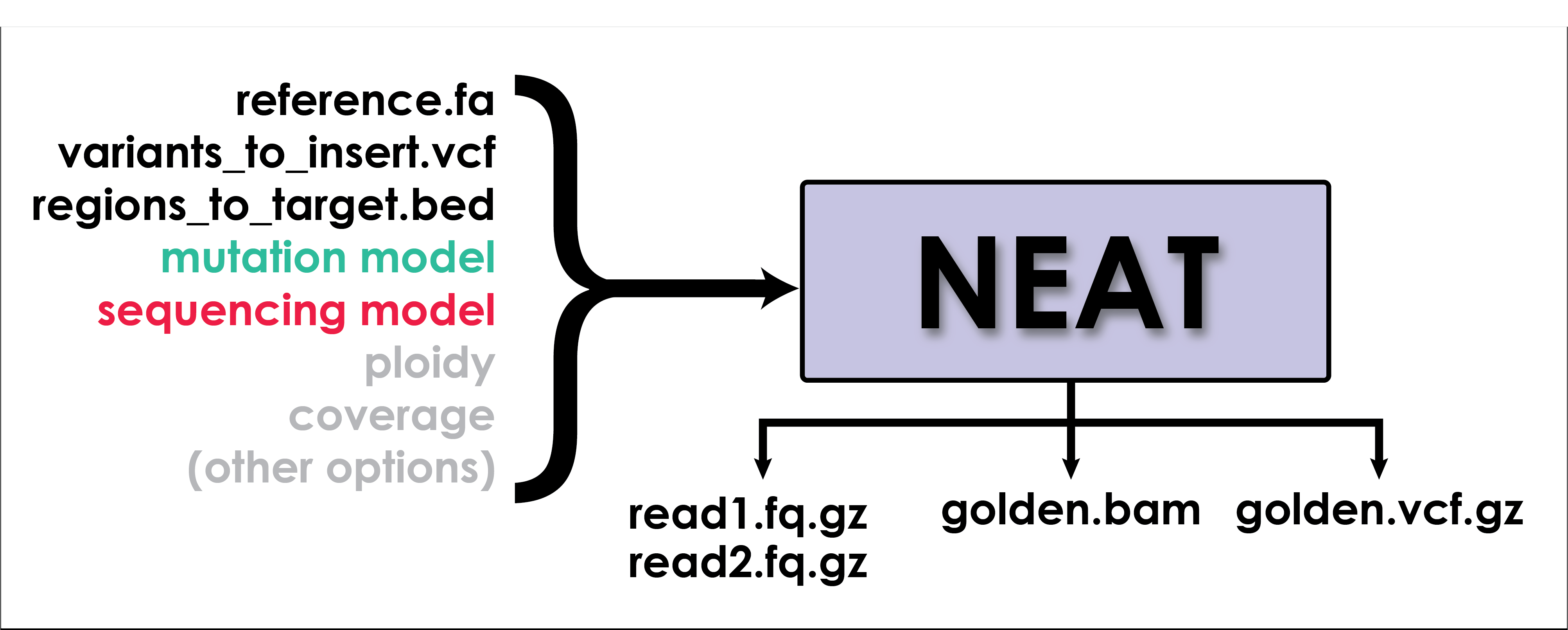
NEAT's gen_reads.py is a fine-grained read simulator. It simulates real-looking data using models learned from specific datasets. There are several supporting utilities for generating models used for simulation and for comparing the outputs of alignment and variant calling to the golden BAM and golden VCF produced by NEAT.
This is an in-progress v3.2 of the software. For a stable release of the previous repo, please see: genReads1 (or check out our v2.0 tag)
To cite this work, please use:
Stephens, Zachary D., Matthew E. Hudson, Liudmila S. Mainzer, Morgan Taschuk, Matthew R. Weber, and Ravishankar K. Iyer. "Simulating next-generation sequencing datasets from empirical mutation and sequencing models." PloS one 11, no. 11 (2016): e0167047.
- python >= 3.8
- biopython == 1.79
- matplotlib >= 3.3.4 (optional, for plotting utilities)
- matplotlib-venn >= 0.11.6 (optional, for plotting utilities)
- pandas >= 1.2.1
- numpy >= 1.22.2
- pysam >= 0.16.0.1
git clone https://github.com/ncsa/NEAT.git
UNTESTED:
python -m pip install -e /path/to/NEAT/
NEAT's core functionality is invoked using the gen_reads.py command. Here's the simplest invocation of gen_reads using default parameters. This command produces a single ended fastq file with reads of length 101, ploidy 2, coverage 10X, using the default sequencing substitution, GC% bias, and mutation rate models.
python gen_reads.py -r sample_data/ecoli.fa -R 101 -o simulated_data
The most commonly added options are --pe (to activate paired-end mode), --bam (to output golden bam), --vcf (to output golden vcf), and -c (to specify average coverage).
| Option | Description |
|---|---|
| -h, --help | Displays usage information |
| -r | Reference sequence file in fasta format. A reference index (.fai) will be created if one is not found in the directory of the reference as [reference filename].fai. Required. The index can be created using samtools faidx. |
| -R | Read length. Required. |
| -o | Output prefix. Use this option to specify where and what to call output files. Required |
| -c | Average coverage across the entire dataset. Default: 10 |
| -e | Sequencing error model data file |
| -E | Average sequencing error rate. The sequencing error rate model is rescaled to make this the average value. |
| -p | Sample Ploidy, default 2 |
| -tr | Bed file containing targeted regions; default coverage for targeted regions is 98% of -c option; default coverage outside targeted regions is 2% of -c option |
| -dr | Bed file with sample regions to discard. |
| -to | off-target coverage scalar [0.02] |
| -m | mutation model data file |
| -M | Average mutation rate. The mutation rate model is rescaled to make this the average value. Must be between 0 and 0.3. These random mutations are inserted in addition to the once specified in the -v option. |
| -Mb | Bed file containing positional mutation rates |
| -N | Below this quality score, base-call's will be replaced with N's |
| -v | Input VCF file. Variants from this VCF will be inserted into the simulated sequence with 100% certainty. |
| --pe | Paired-end fragment length mean and standard deviation. To produce paired end data, one of --pe or --pe-model must be specified. |
| --pe-model | Empirical fragment length distribution. Can be generated using computeFraglen.py. To produce paired end data, one of --pe or --pe-model must be specified. |
| --gc-model | Empirical GC coverage bias distribution. Can be generated using computeGC.py |
| --bam | Output golden BAM file |
| --vcf | Output golden VCF file |
| --fa | Output FASTA instead of FASTQ |
| --rng | rng seed value; identical RNG value should produce identical runs of the program, so things like read locations, variant positions, error positions, etc, should all be the same. |
| --no-fastq | Bypass generation of FASTQ read files |
| --discard-offtarget | Discard reads outside of targeted regions |
| --rescale-qual | Rescale Quality scores to match -E input |
| -d | Turn on debugging mode (useful for development) |
AT produces simulated sequencing datasets. It creates FASTQ files with reads sampled from a provided reference genome, using sequencing error rates and mutation rates learned from real sequencing data. The strength of NEAT lies in the ability for the user to customize many sequencing parameters, produce 'golden,' true positive datasets. We are working on expanding the functionality even further to model more species, generate larger variants, model tumor/normal data and more!
Features:
- Simulate single-end and paired-end reads
- Custom read length
- Can introduce random mutations and/or mutations from a VCF file
- Supported mutation types include SNPs, indels (of any length), inversions, translocations, duplications
- Can emulate multi-ploid heterozygosity for SNPs and small indels
- Can simulate targeted sequencing via BED input specifying regions to sample from
- Can accurately simulate large, single-end reads with high indel error rates (PacBio-like) given a model
- Specify simple fragment length model with mean and standard deviation or an empirically learned fragment distribution using utilities/computeFraglen.py
- Simulates quality scores using either the default model or empirically learned quality scores using utilities/fastq_to_qscoreModel.py
- Introduces sequencing substitution errors using either the default model or empirically learned from utilities/
- Accounts for GC% coverage bias using model learned from utilities/computeGC.py
- Output a VCF file with the 'golden' set of true positive variants. These can be compared to bioinformatics workflow output (includes coverage and allele balance information)
- Output a BAM file with the 'golden' set of aligned reads. These indicate where each read originated and how it should be aligned with the reference
- Create paired tumour/normal datasets using characteristics learned from real tumour data
- Parallelization. COMING SOON!
- Low memory footprint. Constant (proportional to the size of the reference sequence)
The following commands are examples for common types of data to be generated. The simulation uses a reference genome in fasta format to generate reads of 126 bases with default 10X coverage. Outputs paired fastq files, a BAM file and a VCF file. The random variants inserted into the sequence will be present in the VCF and all of the reads will show their proper alignment in the BAM. Unless specified, the simulator will also insert some "sequencing error" -- random variants in some reads that represents false positive results from sequencing.
Simulate whole genome dataset with random variants inserted according to the default model.
python gen_reads.py \
-r hg19.fa \
-R 126 \
-o /home/me/simulated_reads \
--bam \
--vcf \
--pe 300 30
Simulate a targeted region of a genome, e.g. exome, with -tr.
python gen_reads.py \
-r hg19.fa \
-R 126 \
-o /home/me/simulated_reads \
--bam \
--vcf \
--pe 300 30 \
-tr hg19_exome.bed
Simulate a whole genome dataset with only the variants in the provided VCF file using -v and setting mutation rate to 0 with -M.
python gen_reads.py \
-r hg19.fa \
-R 126 \
-o /home/me/simulated_reads \
--bam \
--vcf \
--pe 300 30 \
-v NA12878.vcf \
-M 0
Simulate single-end reads by omitting the --pe option.
python gen_reads.py \
-r hg19.fa \
-R 126 \
-o /home/me/simulated_reads \
--bam \
--vcf
Simulate PacBio-like reads by providing an error model.
python gen_reads.py \
-r hg19.fa \
-R 5000 \
-e models/errorModel_pacbio_toy.pickle.gz \
-E 0.10 \
-o /home/me/simulated_reads
Several scripts are distributed with gen_reads that are used to generate the models used for simulation.
Computes GC% coverage bias distribution from sample (bedrolls genomecov) data. Takes .genomecov files produced by BEDtools genomeCov (with -d option).
bedtools genomecov
-d \
-ibam normal.bam \
-g reference.fa
python compute_gc.py \
-r reference.fa \
-i genomecovfile \
-w [sliding window length] \
-o /path/to/prefix
Computes empirical fragment length distribution from sample data. Takes SAM file via stdin:
python computeFraglen.py \
-i input.bam \
-o /prefix/for/output
and creates fraglen.pickle.gz model in working directory.
Takes references genome and TSV file to generate mutation models:
python gen_mut_model.py \
-r hg19.fa \
-m inputVariants.tsv \
-o /home/me/models
Trinucleotides are identified in the reference genome and the variant file. Frequencies of each trinucleotide transition are calculated and output as a pickle (.p) file.
| Option | Description |
|---|---|
| -r | Reference file for organism in FASTA format. Required |
| -m | Mutation file for organism in VCF format. Required |
| -o | Path to output file and prefix. Required. |
| --bed | Flag that indicates you are using a bed-restricted vcf and fasta (see below) |
| --save-trinuc | Save trinucleotide counts for reference |
| --human-sample | Use to skip unnumbered scaffolds in human references |
| --skip-common | Do not save common snps or high mutation areas |
Generates sequence error model for gen_reads.py -e option. This script needs revision, to improve the quality-score model eventually, and to include code to learn sequencing errors from pileup data.
python genSeqErrorModel.py \
-i input_read1.fq (.gz) / input_read1.sam/bam \
-o /output/prefix \
-i2 input_read2.fq (.gz) / input_read2.sam/bam\
-p input_alignment.pileup \
-q quality score offset [33] \
-Q maximum quality score [41] \
-n maximum number of reads to process [all] \
-s number of simulation iterations [1000000] \
--plot (perform some optional ploting)
Performs plotting and comparison of mutation models generated from genMutModel.py.
python plotMutModel.py \
-i model1.pickle.gz [model2.pickle.gz] [model3.pickle.gz]... \
-l legend_label1 [legend_label2] [legend_label3]... \
-o path/to/pdf_plot_prefix
Tool for comparing VCF files.
python vcf_compare_OLD.py
-r <ref.fa> * Reference Fasta \
-g <golden.vcf> * Golden VCF \
-w <workflow.vcf> * Workflow VCF \
-o <prefix> * Output Prefix \
-m <track.bed> Mappability Track \
-M <int> Maptrack Min Len \
-t <regions.bed> Targetted Regions \
-T <int> Min Region Len \
-c <int> Coverage Filter Threshold [15] \
-a <float> Allele Freq Filter Threshold [0.3] \
--vcf-out Output Match/FN/FP variants [False] \
--no-plot No plotting [False] \
--incl-homs Include homozygous ref calls [False] \
--incl-fail Include calls that failed filters [False] \
--fast No equivalent variant detection [False]
Mappability track examples: https://github.com/zstephens/neat-repeat/tree/master/example_mappabilityTracks
ICGC's "Access Controlled Data" documentation can be found at https://docs.icgc.org/portal/access/. To have access to controlled germline data, a DACO must be submitted. Open tier data can be obtained without a DACO, but germline alleles that do not match the reference genome are masked and replaced with the reference allele. Controlled data includes unmasked germline alleles.