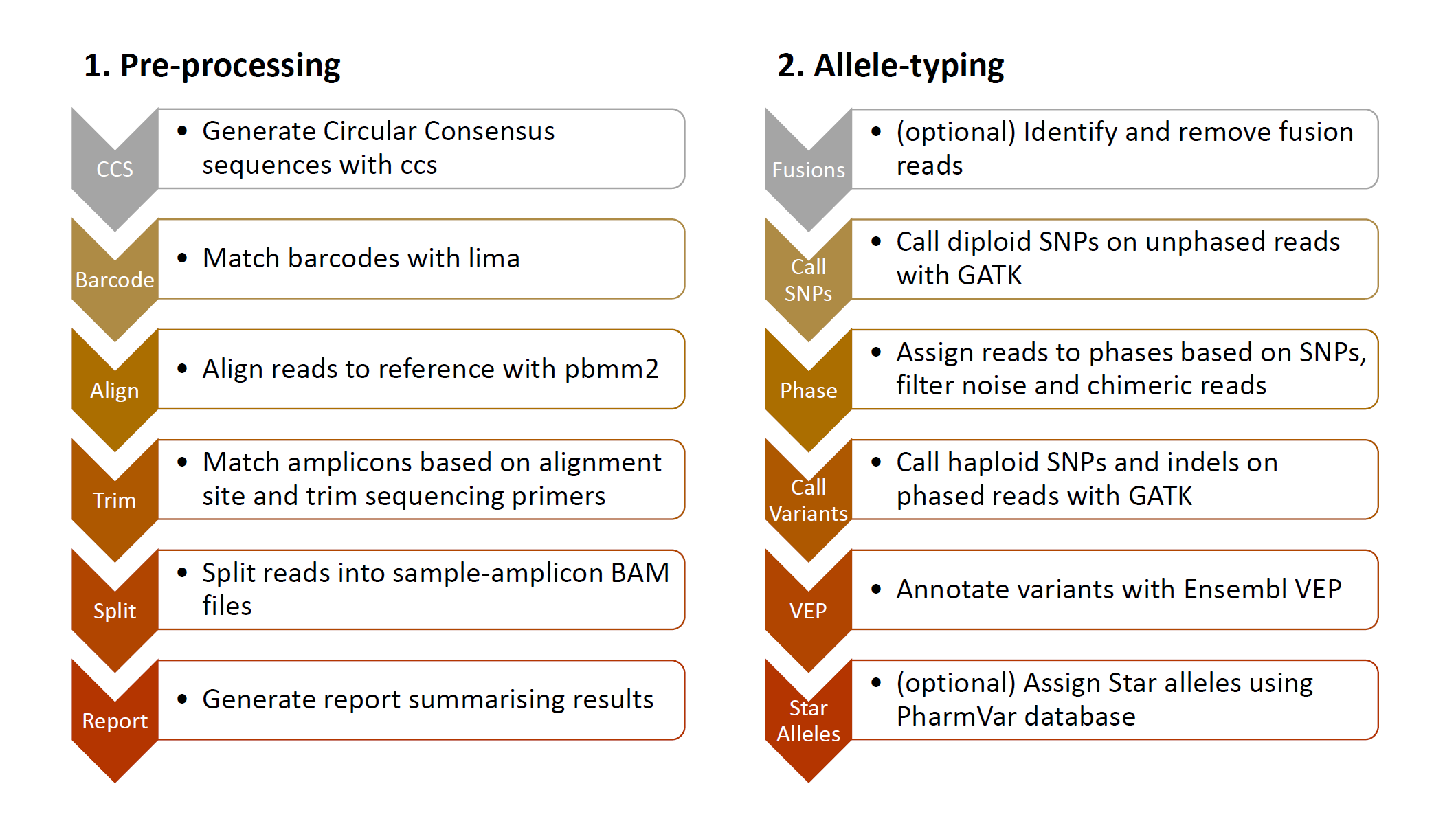
PLASTER is a comprehensive data processing pipeline for allele typing from long amplicon sequencing data generated on the PacBio SMRT platform. Inputs are PacBio subreads in BAM format, as well as sample barcodes and target amplicon details. Outputs are phased BAMs for each sample amplicon and variant calls in VCF format. Additionally the pipeline supports Pharmacogenomic star alelle assignment using the PharmVar database, and gene fusion detection for CYP2D6 and CYP2D7 fusion alleles.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a portable and efficient manner. Included is a Docker container, making installation trivial and results highly reproducible.
- Nextflow (version ≥ 20.07.1)
- Singularity or Docker
- Job compute nodes require an internet connection to download data from PharmVar and Ensembl VEP
-
Running the test dataset
nextflow run bahlolab/PLASTER -profile preproc,test,singularityThis command will download the pipeline from GitHub and run the pre-processing stage on a minimal test dataset using singularity to run the software container. Replace "singularity" with "docker" to use docker instead. Note that nextflow pipelines are run in the current working directory, so make sure your terminal is in the appropriate directory first.
Profiles are provided for executors SLURM and PBS/torque, to use these append either
slurmorpbsto the end of the profile specification (e.g.,-profile preproc,test,singularity,slurm). Additional executors may be specified in a custom nextflow configuration, see Nextflow executor documentation. -
Running your own dataset
nextflow run bahlolab/PLASTER -profile preproc,singularity -c <my_dataset.config>where
my_dataset.configis a Nextflow config file specifying the following required parameters:params { subreads_bam = '/PATH/TO/MY/subreads.bam' barcodes_fasta = '/PATH/TO/MY/barcodes.fasta' amplicons_json = '/PATH/TO/MY/amplicons.json' ref_fasta = 'ftp://hgdownload.cse.ucsc.edu/goldenPath/hg38/chromosomes/chr22.fa.gz' }See Pre-processing Parameters for more details, including details on running in single-sample mode (no barcodes).
-
Resuming a failed run
Adding the-resumeoption to the Nextflow run command will use cached results from any pipeline steps where the inputs remain the same:nextflow run bahlolab/PLASTER -profile preproc,singularity -c <my_dataset.config> -resume -
Outputs
./output/bam/
Directory containing aligned CCS BAM files for each sample-amplicon../output/sample_amplicon_bam_manifest.csv
CSV file with columns "sample", "amplicon", "n_reads", "bam_file" - to be used as input for the Allele-typing stage (see below)../output/pre_processing_report.html
HTML report with various summary statistics and plots.
-
Running the test dataset
nextflow run bahlolab/PLASTER -profile typing,test,singularityThis command will download the pipeline from GitHub and run the allele-typing stage on a minimal test dataset using singularity to run the software container. Replace "singularity" with "docker" to use docker instead. Note that nextflow pipelines are run in the current working directory, so make sure your terminal is in the appropriate directory first.
Profiles are provided for executors SLURM and PBS/torque, to use these append either
slurmorpbsto the end of the profile specification (e.g.,-profile preproc,test,singularity,slurm). Additional executors may be specified in a custom nextflow configuration, see Nextflow executor documentation. -
Running your own dataset
nextflow run bahlolab/PLASTER -profile typing,singularity -c <my_dataset.config>where
my_dataset.configis a file specifying the following required parameters:params { manifest = '/PATH/TO/MY/manifest.csv' amplicons_json = '/PATH/TO/MY/amplicons.json' ref_fasta = 'ftp://hgdownload.cse.ucsc.edu/goldenPath/hg38/chromosomes/chr22.fa.gz' }See Allele-typing Parameters for more details.
-
Resuming a failed run
Adding the-resumeoption to the Nextflow run command will use cached results from any pipeline steps where the inputs remain the same:nextflow run bahlolab/PLASTER -profile preproc,singularity -c <my_dataset.config> -resume -
Outputs
./output/low_read_count.csv
Details of any sample amplicons excluded due to having too few reads. If no sample amplicons are excluded for this reason then this file won't be created../output/low_phased_read_count.csv
Details of any sample amplicons excluded due to having too few phased reads. If no samples amplicons are excluded for this reason then this file won't be created../output/fusion_calls.csv
Details of fusions calls made. Only produced if fusion detection is enabled../output/fusion_report.html
Report summarising fusion calling. Only produced if fusion detection is enabled../output/<amplicon>.phase_copy_num.csv
Details of copy number assigned to each sample-amplicon-phase../output/AmpPhaseR/
Directory containing AmpPhaseR plots showing read phasing, denoising and chimera removal../output/<amplicon>.vep.vcf.gz
VCF file with sample-amplicon-phase calls and VEP variant annotation../output/<amplicon>.sample_phase_alleleles.csv
Star allele assignment for sample phases. Only produced is star allele assignment is enabled../output/<amplicon>.allele_definition.csv
Star allele definitions. Only produced is star allele assignment is enabled.
- CCS - PacificBiosciences/ccs implemented in
pb_ccs.nf - Barcoding - PacificBiosciences/barcoding implemented in
pb_lima.nf - Alignment - PacificBiosciences/pbmm2 implemented in
pb_mm2.nf - Trim - Python script
bam_annotate_amplicons.pybased on pysam - Split - Python scripts
bam_annotate_samples.pyandbam_split_sample_amplicons.pybased on pysam - Report - Rmarkdown document
preproc-report.Rmd
- Fusions - R script
fusion_call.Rand Rmarkdown documentfusion_report.Rmd - Call SNPs - GATK HaplotypeCaller and GATK GenotypeGVCFs implemented in
gatk.nf - Phase - BCFtools and R package AmpPhaseR implemented in
phase.nf - Call Variants - GATK HaplotypeCaller and GATK GenotypeGVCFs implemented in
gatk.nf - VEP - Ensembl Variant Effect Predictor implemented in
vep.nf - Star Alleles - R script
pharmvar_star_allele.Rusing PharmVar database
- Contributions are welcome, feel free to fork this repository and make a pull request.
- The pipeline could be extended to support Nanopore data as well PacBio data, but would require an alternate pre-processing stage. Please contact us if you are interested in working on this.
- The folder
CopyNumAnalysiscontains raw data and the analysis Rmarkdown document associated with the qPCR copy number analysis presented in the PLASTER manuscript.