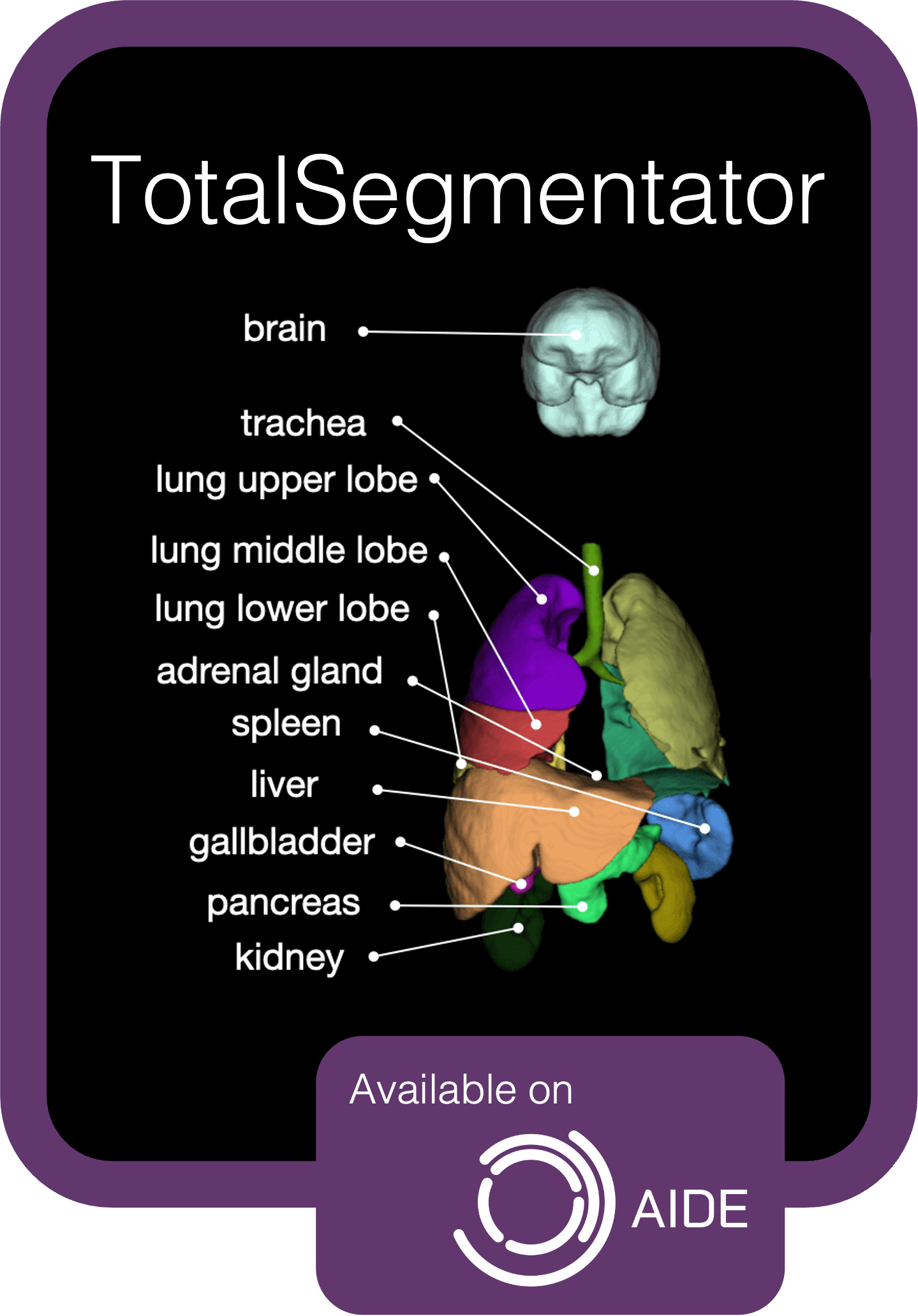
TotalSegmentator packaged as an AIDE Application, based on
the open-source
MONAI Application Package (MAP) standard.
View repo
·
Report Bug
·
Request Feature
The input to the TotalSegmentator MAP is a folder containing CT DICOM .dcm files. The output is an RT Struct DICOM .dcm
file containing the contours of all the segmented regions.
The TotalSegmentator MAP consists of three operators:
dcm2nii_operator.py– converts the input CT DICOM files to NIfTI format required by TotalSegmentator, using dcm2niixtotalsegmentator_operator.py– runs TotalSegmentatorrtstructwriter_operator.py– converts the output segmentations from NIfTI to DICOM RT Struct format, using rt-utilsclinrev_pdf_operator.py– creates a DICOM encapsulated PDF showing three views of the input CT dataset overlaid with the segmentations generated by TotalSegmentator
- GPU-enabled machine
- Docker
- Required for running the MAP. Not required for running source code
- Download
git clone https://github.com/GSTT-CSC/TotalSegmentator-AIDE.git- Setup virtual env
cd TotalSegmentator-AIDE
python -m venv venv
source venv/bin/activate
pip install --upgrade pip setuptools wheel
pip install -r requirements.txt- Create
inputandoutputdirectories
mkdir input output- Ensure Python venv running
- Copy DICOM
.dcmfiles toinput/directory
monai-deploy exec app -i input/ -o -output/
# alternatively:
python app -i input/ -o output/- Ensure Python venv running
- Ensure Docker running
- Copy DICOM
.dcmfiles toinput/directory
Important: we create an initial MAP map-init upon which we build any 3rd-party non-Python software (e.g. dcm2niix).
The final MAP is called map
# Initial packaging of MAP
monai-deploy package app --tag ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0 -l DEBUG
# Build 3rd-party software on top of MAP
docker build -t ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 app/
# Test MAP with MONAI Deploy
monai-deploy run ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 input/ output/
# Push initial MAP and final MAP to GHCR
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0Enter Docker container for testing
docker run --gpus all -it --rm -v /TotalSegmentator-AIDE/input:/var/monai/input/ --entrypoint /bin/bash ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0Run on specified GPU if machine has >1 available
CUDA_VISIBLE_DEVICES=2 monai-deploy run ghcr.io/gstt-csc/totalsegmentator-aide/map:0.2.0 input/ output/Reminder: if GHCR requires Personal Access Token
echo $CR_PAT | docker login ghcr.io -u <github-username> --password-stdin
docker push ghcr.io/gstt-csc/totalsegmentator-aide/map-init:0.2.0Note: two containers are present under GitHub Packages:
The map version is for general usage and deployment. The map-init version is for developers who wish to build the
MAP from scratch as part of the two-step build process described above.
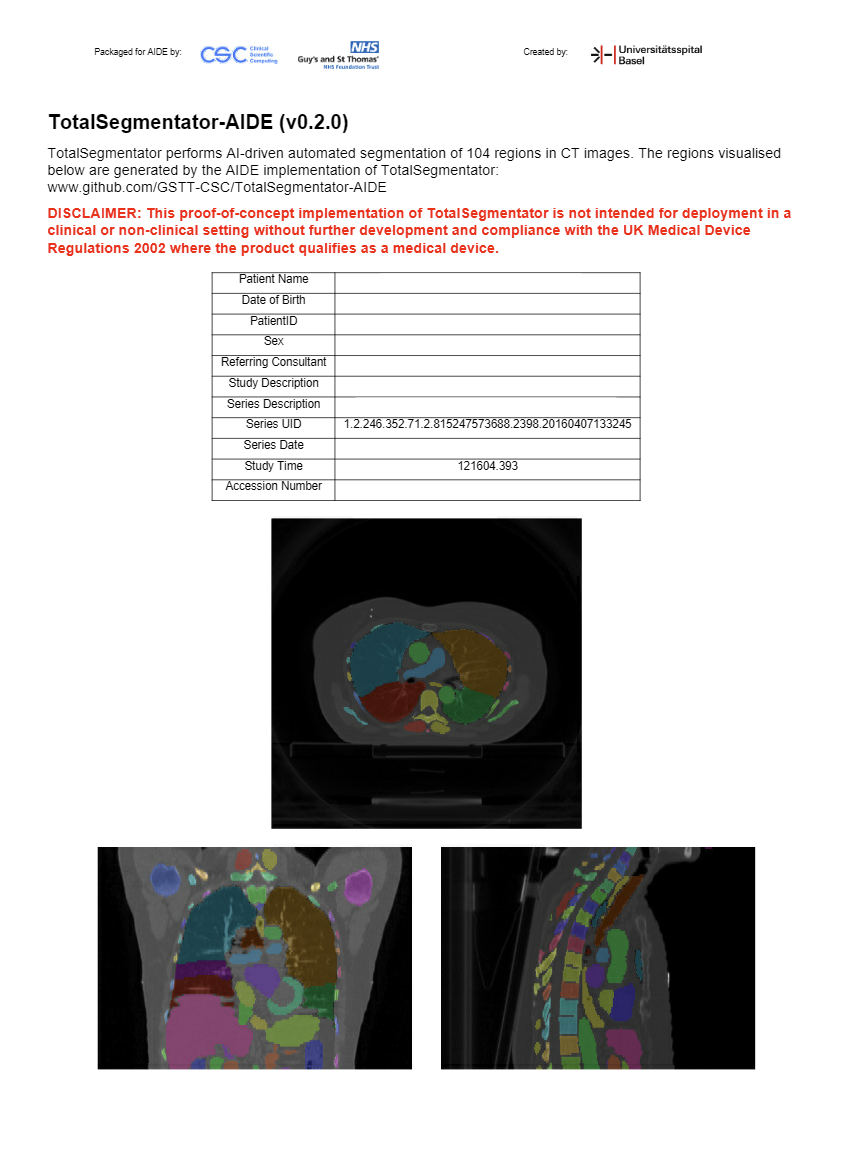
AIDE Clinical Review is a user interface within AIDE for assessing the quality of AI results. It can display DICOM
Encapsulated PDFs. The screenshot below is an example PDF generated by clinrev_pdf_operator.py:
To run TotalSegmentator on AIDE, two files from the app/workflows directory are required, namely:
totalsegmentator.json– AIDE Clinical Workflow file.- Effectively a sequence of tasks run on AIDE
totalsegmentator-argo-template.yaml– Argo Workflow Template file.- Called by the AIDE Clinical Workflow file. Executes the Totalsegmentator-AIDE MAP using Argo. This is the central task within the AIDE Clinical Workflow file.
The app/workflows directory also contains four illustrative Argo Workflow Templates depending on system performance.
low, medium, high and dgx examples are provided with different CPU and RAM settings.