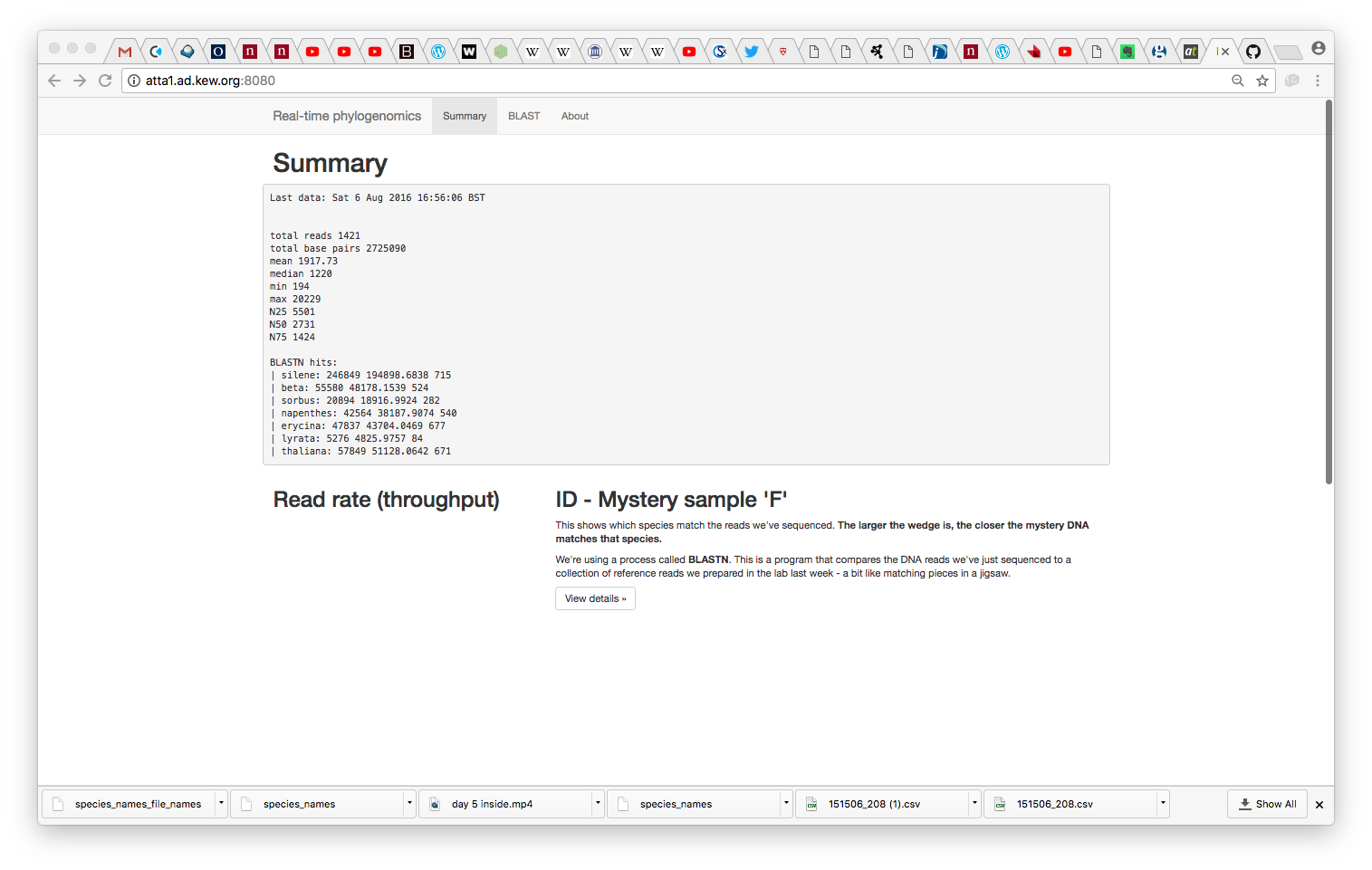
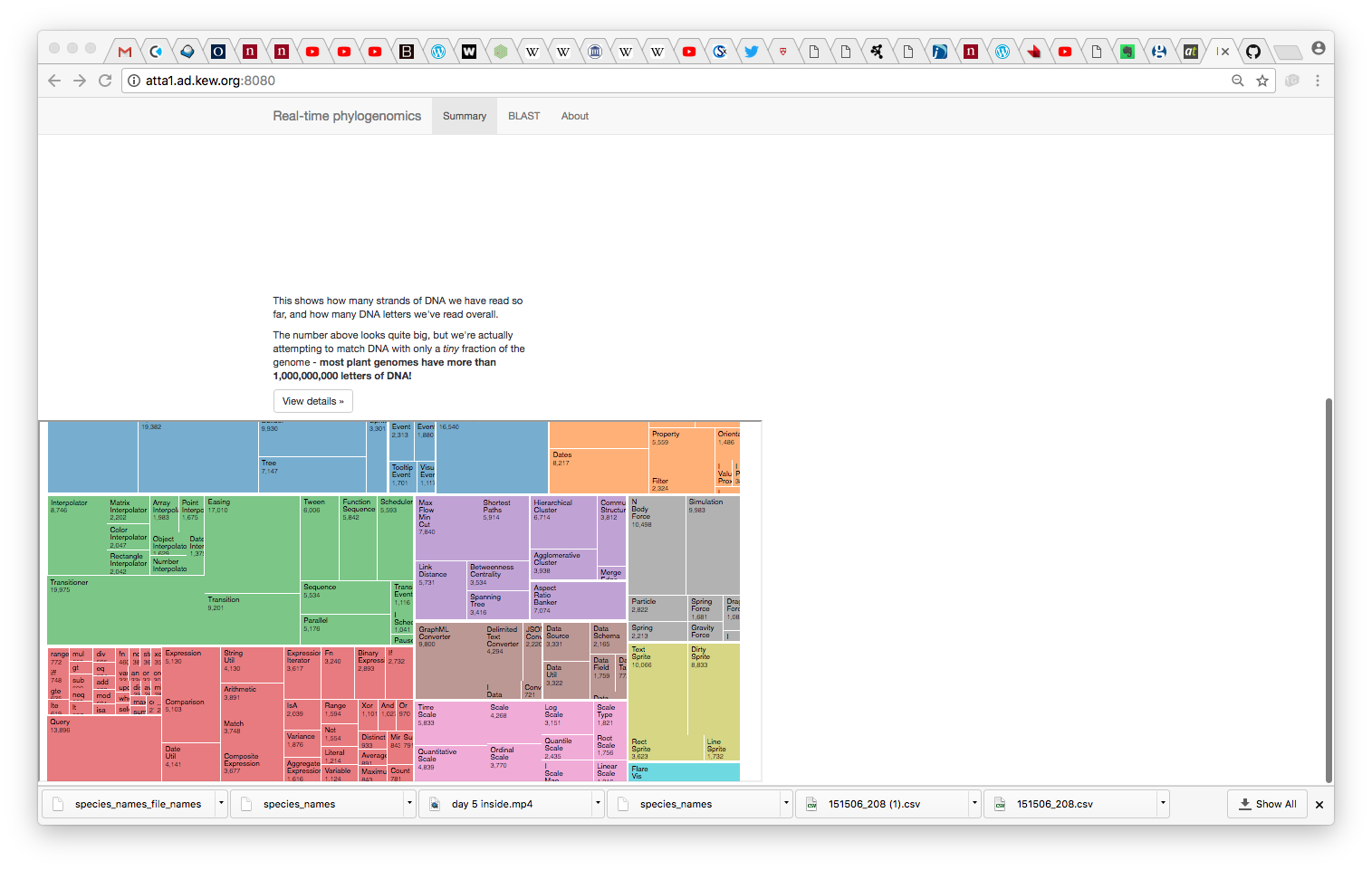
Getting a Taxonomy Treemap streaming from a MinION device
We wanted to come up with a real-time platform to analyse (classify) and visualise a metagenome against ~500 fungal genomes.
This consists of:
- A subset of the NCBI taxonomy pulled by
horrible_ncbi_taxonomy/ncbi_ids.Rmdwhich attempts to get a taxonomy (proxy for phylogeny) for the JGI fungal genomes - A directory watcher (
SimpleDirWatcher.jar) which triggers a bash script (do_metahack.sh) when new reads appear in the/inputdirectory - An analysis pipeline forked from
do_metahack.shwhich (at present) BLASTs reads against the 500 (actually 300) genome database - A D3-based visualiser which uses a treemap paradigm to classify reads and cascade confidence information up the taxonomy to aggregate ID information by genus/family/phylum
Sequence reads in FASTA
Served to html:
There's a google doc with more but:
- Programatically pull taxonomy more flexibly
- Debug D3 tree expansion
- Refine actual ID engine