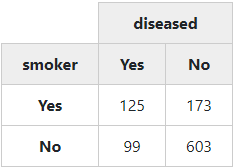The following vignettes complement this page:
Recommendations for Using summarytools With
Rmarkdown
Introduction to
summarytools
– Contents similar to this page (minus installation instructions), with
fancier table stylings.
summarytools is an R package providing tools to neatly and quickly summarize data. It can also make R a little easier to learn and to use, especially for data cleaning and preliminary analysis. Four functions are at the core of the package:
freq(): frequency tables with proportions, cumulative proportions and missing data informationctable(): cross-tabulations between two factors or any discrete data, with total, rows or columns proportions, as well as marginal totalsdescr(): descriptive (univariate) statistics for numerical datadfSummary(): Extensive data frame summaries that facilitate data cleaning and firsthand evaluation
An emphasis has been put on both what and how results are presented, so that the package can serve both as an exploration and reporting tool, used on its own for minimal reports, or with other sets of tools such as rmarkdown, and knitr.
Building on the strengths of pander and htmltools, the outputs produced by summarytools can be:
- Displayed in plain text in the R console (default behaviour)
- Used in Rmarkdown documents and knitted along with other text and R output
- Written to html files that fire up in RStudio’s Viewer pane or in the default browser
- Written to plain or Rmarkdown text files
It is also possible to include summarytools’ functions in Shiny apps.
Version 0.9 brought many changes and improvements. A summary of those changes can be found near the end of this page. Changes specific to the latest release can be found in the NEWS file.
This is the recommended method, as some minor fixes are made available between CRAN releases.
Magick++ Dependency on Linux and Mac OS
Before proceeding, you must install Magick++
- deb: 'libmagick++-dev' (Debian, Ubuntu)
- rpm: 'ImageMagick-c++-devel' (Fedora, CentOS, RHEL)
- csw: 'imagemagick_dev' (Solaris)
On MacOS it is recommended to use install ImageMagick-6 from homebrew
with extra support for fontconfig and rsvg rendering:
brew reinstall imagemagick@6 --with-fontconfig --with-librsvg
For older Ubuntu versions Trusty (14.04) and Xenial (16.04) use the PPA:
sudo add-apt-repository -y ppa:opencpu/imagemagick
sudo apt-get update
sudo apt-get install -y libmagick++-dev
After this is done, proceed with the installation:
install.packages("devtools")
library(devtools)
install_github("rapporter/pander") # Necessary for optimal results!
install_github("dcomtois/summarytools")Simply install it with install.packages():
install.packages("summarytools")The official documentation can be found here.
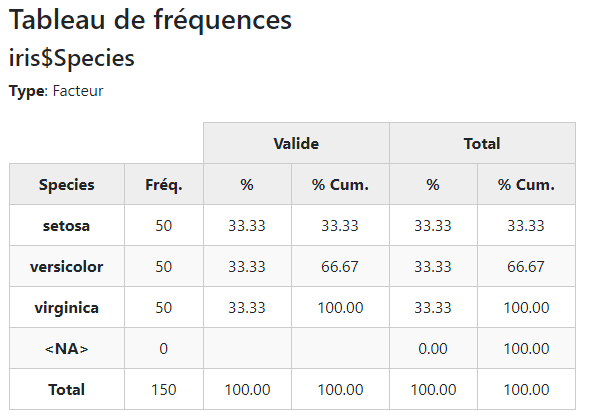
The freq() function generates a table of frequencies with counts and
proportions. Since GitHub uses markdown rendering, we’ve set the
style argument to “rmarkdown”. When creating Rmd documents,
knitr takes care of converting the generated markup characters into
actual html.
library(summarytools)
freq(iris$Species, style = "rmarkdown")iris$Species
Type:
Factor
| Freq | % Valid | % Valid Cum. | % Total | % Total Cum. | |
|---|---|---|---|---|---|
| setosa | 50 | 33.33 | 33.33 | 33.33 | 33.33 |
| versicolor | 50 | 33.33 | 66.67 | 33.33 | 66.67 |
| virginica | 50 | 33.33 | 100.00 | 33.33 | 100.00 |
| <NA> | 0 | 0.00 | 100.00 | ||
| Total | 150 | 100.00 | 100.00 | 100.00 | 100.00 |
If we do not worry about missing data, we can set report.nas = FALSE:
freq(iris$Species, report.nas = FALSE, style = "rmarkdown", headings = FALSE)| Freq | % | % Cum. | |
|---|---|---|---|
| setosa | 50 | 33.33 | 33.33 |
| versicolor | 50 | 33.33 | 66.67 |
| virginica | 50 | 33.33 | 100.00 |
| Total | 150 | 100.00 | 100.00 |
We can simplify the results further and omit the Totals row by
specifying totals = FALSE, as well as omit the cumulative rows by
setting cumul = FALSE.
freq(iris$Species, report.nas = FALSE, totals = FALSE, cumul = FALSE, style = "rmarkdown", headings = FALSE)| Freq | % | |
|---|---|---|
| setosa | 50 | 33.33 |
| versicolor | 50 | 33.33 |
| virginica | 50 | 33.33 |
To get familiar with the various output styles, try different values for
style – “simple”, “rmarkdown” or “grid”, and see how this affects the
results in the console.
The “rows” argument allows subsetting the resulting frequency table; we can use it in 3 different ways:
- To select rows by position, we use a numerical vector;
rows = 1:10will show the frequencies for the first 10 values only - To select rows by name, we either use
- a character vector specifying all desired values (row names)
- a single character string to be used as a regular expression; only the matching values will be displayed
Used in combination with the “order” argument, this can be quite
practical. Say we have a character variable containing many distinct
values and wish to know which ones are the 10 most frequent. To achieve
this, we would simply use order = "freq" along with rows = 1:5.
There is more than one way to do this, but the best approach is to
simply pass the data frame object (subsetted if needed) to freq():
(results not shown)
freq(tobacco[ ,c("gender", "age.gr", "smoker")])We can without fear pass a whole data frame to freq(); it will figure
out which variables to ignore (numerical variables having many distinct
values).
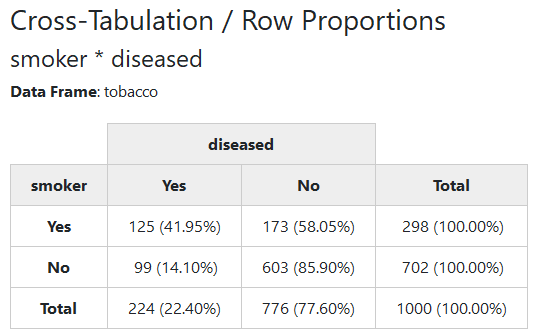
We’ll now use a sample data frame called tobacco, which is included in
summarytools. We want to cross-tabulate two categorical variables:
smoker and diseased.
Since markdown does not support multiline headings, we’ll show a rendered html version of the results:
print(ctable(tobacco$smoker, tobacco$diseased, prop = "r"), method = "render")Note that we have to set the knitr chunk option results to “asis”
for the results to appear as they should.
By default, ctable() shows row proportions. To show column or total
proportions, use prop = "c" or prop = "t", respectively. To omit
proportions, use prop = "n".
In the next example, we’ll create a simple “2 x 2” table (no proportions, no totals):
with(tobacco,
print(ctable(smoker, diseased, prop = 'n', totals = FALSE),
headings = FALSE, method = "render"))To display chi-square results below the table, set the “chisq” parameter
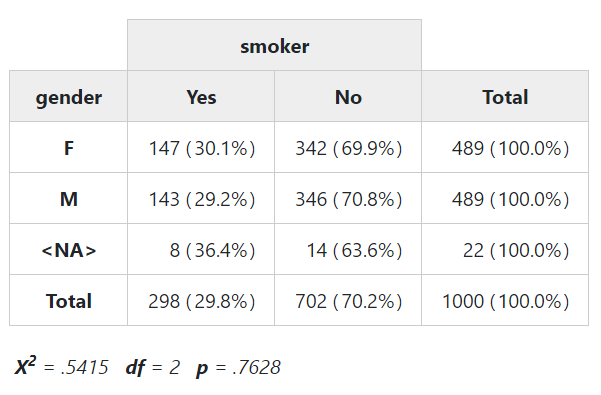
to TRUE. This time, instead of with(), we’ll use the %$% operator
from the magrittr package, which works in a very similar fashion.
library(magrittr)
tobacco %$% ctable(gender, smoker, chisq = TRUE, headings = FALSE)Note that a warning will be issued when at least one expected cell counts is lower than 5.
The descr() function generates common central tendency statistics and
measures of dispersion for numerical data. It can handle single vectors
as well as data frames, in which case it will ignore non-numerical
columns (and display a message to that effect).
descr(iris, style = "rmarkdown")iris
N:
150
| Petal.Length | Petal.Width | Sepal.Length | Sepal.Width | |
|---|---|---|---|---|
| Mean | 3.76 | 1.20 | 5.84 | 3.06 |
| Std.Dev | 1.77 | 0.76 | 0.83 | 0.44 |
| Min | 1.00 | 0.10 | 4.30 | 2.00 |
| Q1 | 1.60 | 0.30 | 5.10 | 2.80 |
| Median | 4.35 | 1.30 | 5.80 | 3.00 |
| Q3 | 5.10 | 1.80 | 6.40 | 3.30 |
| Max | 6.90 | 2.50 | 7.90 | 4.40 |
| MAD | 1.85 | 1.04 | 1.04 | 0.44 |
| IQR | 3.50 | 1.50 | 1.30 | 0.50 |
| CV | 0.47 | 0.64 | 0.14 | 0.14 |
| Skewness | -0.27 | -0.10 | 0.31 | 0.31 |
| SE.Skewness | 0.20 | 0.20 | 0.20 | 0.20 |
| Kurtosis | -1.42 | -1.36 | -0.61 | 0.14 |
| N.Valid | 150.00 | 150.00 | 150.00 | 150.00 |
| Pct.Valid | 100.00 | 100.00 | 100.00 | 100.00 |
If your eyes/brain prefer seeing things the other way around, just use
transpose = TRUE. Here, we also select only the statistics we wish to
see, and specify headings = FALSE to avoid reprinting the same
information as above.
We specify the stats we wish to report with the stats argument, which
also accepts values “all”, “fivenum”, and “common”. See ?descr for a
complete list of available statistics.
descr(iris, stats = "common", transpose = TRUE, headings = FALSE, style = "rmarkdown")| Mean | Std.Dev | Min | Median | Max | N.Valid | Pct.Valid | |
|---|---|---|---|---|---|---|---|
| Petal.Length | 3.76 | 1.77 | 1.00 | 4.35 | 6.90 | 150.00 | 100.00 |
| Petal.Width | 1.20 | 0.76 | 0.10 | 1.30 | 2.50 | 150.00 | 100.00 |
| Sepal.Length | 5.84 | 0.83 | 4.30 | 5.80 | 7.90 | 150.00 | 100.00 |
| Sepal.Width | 3.06 | 0.44 | 2.00 | 3.00 | 4.40 | 150.00 | 100.00 |
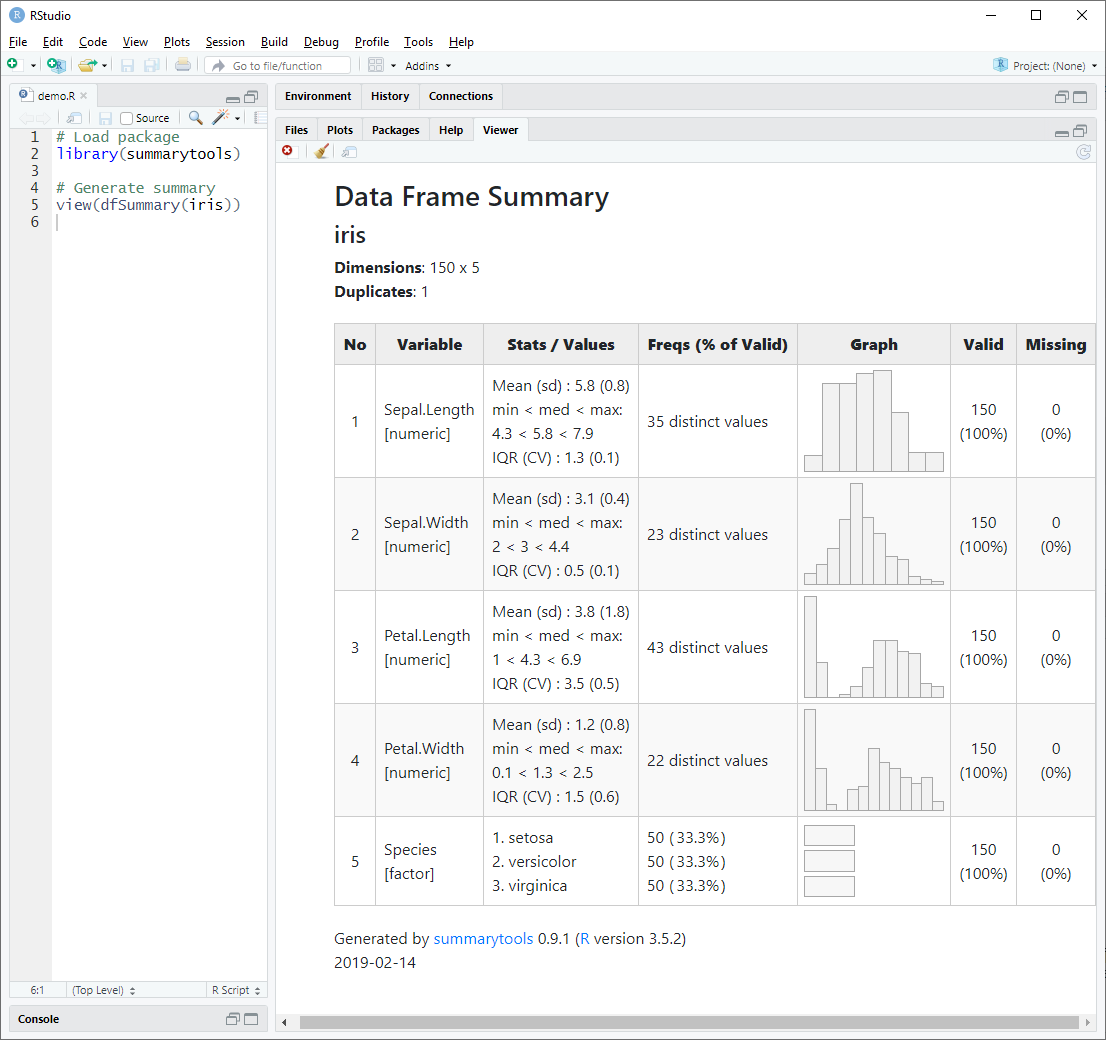
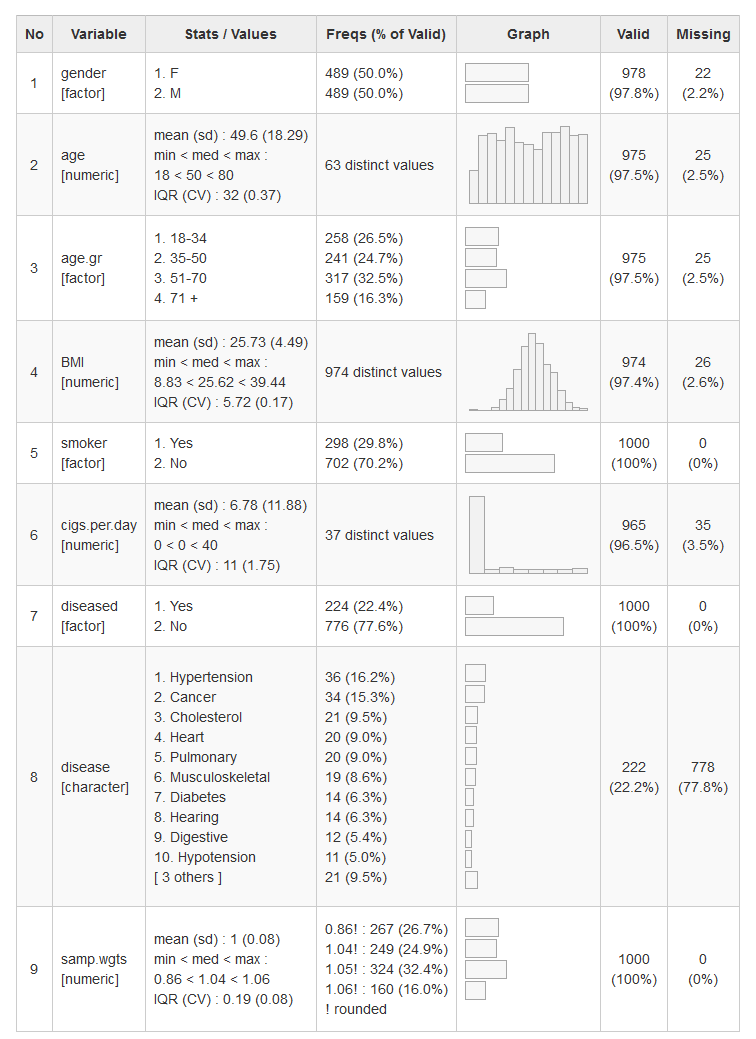
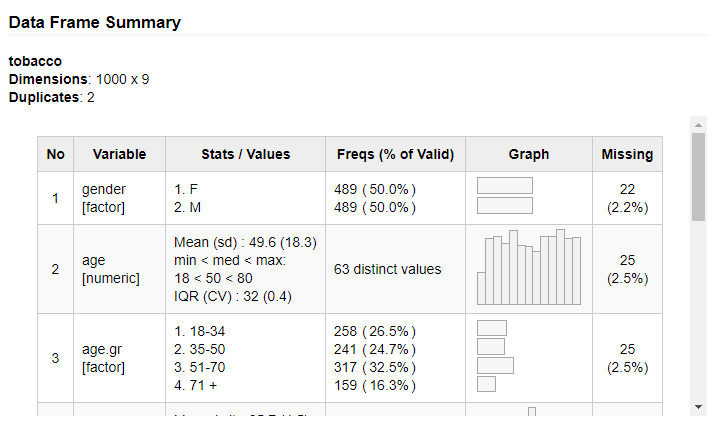
dfSummary() collects information about all variables in a data frame
and displays it in a single legible table.
To generate a summary report and have it displayed in RStudio’s Viewer pane (or in the default Web browser if working outside RStudio), we simply do as follows:
library(summarytools)
view(dfSummary(iris))Of course, it is also possible to use dfSummary() in Rmarkdown
documents. It is usually a good idea to exclude a column or two,
otherwise the table might be a bit too wide. For instance, since the
Valid and NA columns are redundant, we can drop one of them.
dfSummary(tobacco, plain.ascii = FALSE, style = "grid",
graph.magnif = 0.75, valid.col = FALSE, tmp.img.dir = "/tmp")While rendering html tables with view() doesn’t require it, here it
is essential to specify tmp.img.dir. We’ll explain why further
below.
When generating freq() or descr() tables, it is possible to turn the
results into “tidy” tables with the use of the tb() function (think of
tb as a diminutive for tibble). For example:
library(magrittr)
iris %>% descr(stats = "common") %>% tb()## # A tibble: 4 x 8
## variable mean sd min med max n.valid pct.valid
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Petal.Length 3.76 1.77 1 4.35 6.9 150 100
## 2 Petal.Width 1.20 0.762 0.1 1.3 2.5 150 100
## 3 Sepal.Length 5.84 0.828 4.3 5.8 7.9 150 100
## 4 Sepal.Width 3.06 0.436 2 3 4.4 150 100
iris$Species %>% freq(cumul = FALSE, report.nas = FALSE) %>% tb()## # A tibble: 3 x 3
## value freq pct
## <fct> <dbl> <dbl>
## 1 setosa 50 33.3
## 2 versicolor 50 33.3
## 3 virginica 50 33.3
By definition, no total rows are part of tidy tables, and row.names are converted to regular columns.
Here are two examples of how lists created using stby() are
transformed into tibbles. Notice how the order parameter affects the
table’s row ordering:
grouped_freqs <- stby(data = tobacco$smoker, INDICES = tobacco$gender,
FUN = freq, cumul = FALSE, report.nas = FALSE)
grouped_freqs %>% tb()## # A tibble: 4 x 4
## gender smoker freq pct
## <fct> <fct> <dbl> <dbl>
## 1 F Yes 147 15.0
## 2 F No 342 35.0
## 3 M Yes 143 14.6
## 4 M No 346 35.4
grouped_freqs %>% tb(order = 2)## # A tibble: 4 x 4
## gender smoker freq pct
## <fct> <fct> <dbl> <dbl>
## 1 F Yes 147 15.0
## 2 M Yes 143 14.6
## 3 F No 342 35.0
## 4 M No 346 35.4
grouped_descr <- stby(data = exams, INDICES = exams$gender,
FUN = descr, stats = "common")
grouped_descr %>% tb()## # A tibble: 12 x 9
## gender variable mean sd min med max n.valid pct.valid
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Girl economics 72.5 7.79 62.3 70.2 89.6 14 93.3
## 2 Girl english 73.9 9.41 58.3 71.8 93.1 14 93.3
## 3 Girl french 71.1 12.4 44.8 68.4 93.7 14 93.3
## 4 Girl geography 67.3 8.26 50.4 67.3 78.9 15 100
## 5 Girl history 71.2 9.17 53.9 72.9 86.4 15 100
## 6 Girl math 73.8 9.03 55.6 74.8 86.3 14 93.3
## 7 Boy economics 75.2 9.40 60.5 71.7 94.2 15 100
## 8 Boy english 77.8 5.94 69.6 77.6 90.2 15 100
## 9 Boy french 76.6 8.63 63.2 74.8 94.7 15 100
## 10 Boy geography 73 12.4 47.2 71.2 96.3 14 93.3
## 11 Boy history 74.4 11.2 54.4 72.6 93.5 15 100
## 12 Boy math 73.3 9.68 60.5 72.2 93.2 14 93.3
grouped_descr %>% tb(order = 2)## # A tibble: 12 x 9
## gender variable mean sd min med max n.valid pct.valid
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Girl economics 72.5 7.79 62.3 70.2 89.6 14 93.3
## 2 Boy economics 75.2 9.40 60.5 71.7 94.2 15 100
## 3 Girl english 73.9 9.41 58.3 71.8 93.1 14 93.3
## 4 Boy english 77.8 5.94 69.6 77.6 90.2 15 100
## 5 Girl french 71.1 12.4 44.8 68.4 93.7 14 93.3
## 6 Boy french 76.6 8.63 63.2 74.8 94.7 15 100
## 7 Girl geography 67.3 8.26 50.4 67.3 78.9 15 100
## 8 Boy geography 73 12.4 47.2 71.2 96.3 14 93.3
## 9 Girl history 71.2 9.17 53.9 72.9 86.4 15 100
## 10 Boy history 74.4 11.2 54.4 72.6 93.5 15 100
## 11 Girl math 73.8 9.03 55.6 74.8 86.3 14 93.3
## 12 Boy math 73.3 9.68 60.5 72.2 93.2 14 93.3
summarytools has a generic print method, print.summarytools().
By default, its method argument is set to “pander”. One of the ways in
which view() is useful is that we can use it to easily display html
outputs in RStudio’s Viewer. The view() function simply acts as a
wrapper around print.summarytools(), specifying method = 'viewer'.
When used outside RStudio, method falls back to “browser” and the
report is shown in the system’s default browser.
We can use stby() the same way as R’s base function by() with the
four core summarytools functions. This returns a list-type object
containing as many elements as there are categories in the grouping
variable.
Why not just use by()? The reason is that by() creates objects
of class “by()”, which have a dedicated print() method conflicting
with summarytools’ way of printing list-type objects. Since print.by()
can’t be redefined (as of CRAN policies), the sensible solution was to
introduce a function that is essentially a clone of by(), except that
the objects it creates have the class “stby”, allowing the desired
flexibility.
Using the iris data frame, we will now display descriptive statistics by Species.
(iris_stats_by_species <- stby(data = iris,
INDICES = iris$Species,
FUN = descr, stats = c("mean", "sd", "min", "med", "max"),
transpose = TRUE))## Non-numerical variable(s) ignored: Species
iris
Group: Species = setosa
N: 50
| Mean | Std.Dev | Min | Median | Max | |
|---|---|---|---|---|---|
| Petal.Length | 1.46 | 0.17 | 1.00 | 1.50 | 1.90 |
| Petal.Width | 0.25 | 0.11 | 0.10 | 0.20 | 0.60 |
| Sepal.Length | 5.01 | 0.35 | 4.30 | 5.00 | 5.80 |
| Sepal.Width | 3.43 | 0.38 | 2.30 | 3.40 | 4.40 |
Group: Species = versicolor
N: 50
| Mean | Std.Dev | Min | Median | Max | |
|---|---|---|---|---|---|
| Petal.Length | 4.26 | 0.47 | 3.00 | 4.35 | 5.10 |
| Petal.Width | 1.33 | 0.20 | 1.00 | 1.30 | 1.80 |
| Sepal.Length | 5.94 | 0.52 | 4.90 | 5.90 | 7.00 |
| Sepal.Width | 2.77 | 0.31 | 2.00 | 2.80 | 3.40 |
Group: Species = virginica
N: 50
| Mean | Std.Dev | Min | Median | Max | |
|---|---|---|---|---|---|
| Petal.Length | 5.55 | 0.55 | 4.50 | 5.55 | 6.90 |
| Petal.Width | 2.03 | 0.27 | 1.40 | 2.00 | 2.50 |
| Sepal.Length | 6.59 | 0.64 | 4.90 | 6.50 | 7.90 |
| Sepal.Width | 2.97 | 0.32 | 2.20 | 3.00 | 3.80 |
To see an html version of these results, we simply use view() (also
possible is to use print() with method = "viewer"): (results not
shown)
view(iris_stats_by_species)
# or
print(iris_stats_by_species, method = "viewer")A special situation occurs when we want grouped statistics for one variable only. Instead of showing several tables, each having one column, summarytools assembles everything into a single table:
data(tobacco)
with(tobacco, stby(BMI, age.gr, descr,
stats = c("mean", "sd", "min", "med", "max")))BMI by age.gr
Data Frame: tobacco
N: 258
| 18-34 | 35-50 | 51-70 | 71 + | |
|---|---|---|---|---|
| Mean | 23.84 | 25.11 | 26.91 | 27.45 |
| Std.Dev | 4.23 | 4.34 | 4.26 | 4.37 |
| Min | 8.83 | 10.35 | 9.01 | 16.36 |
| Median | 24.04 | 25.11 | 26.77 | 27.52 |
| Max | 34.84 | 39.44 | 39.21 | 38.37 |
The transposed version looks like this:
| Mean | Std.Dev | Min | Median | Max | |
|---|---|---|---|---|---|
| 18-34 | 23.84 | 4.23 | 8.83 | 24.04 | 34.84 |
| 35-50 | 25.11 | 4.34 | 10.35 | 25.11 | 39.44 |
| 51-70 | 26.91 | 4.26 | 9.01 | 26.77 | 39.21 |
| 71 + | 27.45 | 4.37 | 16.36 | 27.52 | 38.37 |
This is a little trickier – the working syntax is as follows:
stby(list(x = tobacco$smoker, y = tobacco$diseased), tobacco$gender, ctable)
# or equivalently
with(tobacco, stby(list(x = smoker, y = diseased), gender, ctable))To create grouped statistics with descr() or dfSummary(), it is
possible to use dplyr’s group_by() as an alternative to stby().
Aside from the syntactic differences, one key distinction is that
dplyr::group_by() considers NA values on the grouping variables as
valid categories, albeit with a warning message suggesting to use
forcats::fct_explicit_na to make NA’s explicit. The best way to go
is simply to follow that advice:
library(dplyr)
tobacco$gender <- forcats::fct_explicit_na(tobacco$gender)
tobacco %>% group_by(gender) %>% descr(stats = "fivenum")## Non-numerical variable(s) ignored: gender, age.gr, smoker, diseased, disease
tobacco
Group: gender = F
N: 489
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 18.00 | 9.01 | 0.00 | 0.86 |
| Q1 | 34.00 | 22.98 | 0.00 | 0.86 |
| Median | 50.00 | 25.87 | 0.00 | 1.04 |
| Q3 | 66.00 | 29.48 | 10.50 | 1.05 |
| Max | 80.00 | 39.44 | 40.00 | 1.06 |
Group: gender = M
N: 489
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 18.00 | 8.83 | 0.00 | 0.86 |
| Q1 | 34.00 | 22.52 | 0.00 | 0.86 |
| Median | 49.50 | 25.14 | 0.00 | 1.04 |
| Q3 | 66.00 | 27.96 | 11.00 | 1.05 |
| Max | 80.00 | 36.76 | 40.00 | 1.06 |
Group: gender = (Missing)
N: 22
| age | BMI | cigs.per.day | samp.wgts | |
|---|---|---|---|---|
| Min | 19.00 | 20.24 | 0.00 | 0.86 |
| Q1 | 36.00 | 24.97 | 0.00 | 1.04 |
| Median | 55.50 | 27.16 | 0.00 | 1.05 |
| Q3 | 64.00 | 30.23 | 10.00 | 1.05 |
| Max | 80.00 | 32.43 | 28.00 | 1.06 |
As we have seen, summarytools can generate both text/markdown and html results. Both types of outputs can be used in Rmarkdown documents. The vignette Recommendations for Using summarytools With Rmarkdown provides good guidelines, but here are a few tips to get started:
- Always set the
knitrchunk optionresults = 'asis'. You can do this on a chunk-by-chunk basis, but it is easier to just set it globally in a “setup” chunk:
knitr::opts_chunk$set(echo = TRUE, results = 'asis')Refer to this page to learn more about knitr’s options.
- To get better results when generating html output with
method = 'render', set up your .Rmd document so that it includes summarytools’ css. Thest_css()function makes this very easy.
# ---
# title: "RMarkdown using summarytools"
# output: html_document
# ---
#
# ```{r setup, include=FALSE}
# library(knitr)
# opts_chunk$set(comment = NA, prompt = FALSE, cache = FALSE, results = 'asis')
# library(summarytools)
# st_options(plain.ascii = FALSE, # This is a must in Rmd documents
# style = "rmarkdown", # idem
# dfSummary.varnumbers = FALSE, # This keeps results narrow enough
# dfSummary.valid.col = FALSE) # idem
#```
#
# ```{r, echo=FALSE}
# st_css()
# ```
Since results = 'asis' can conflict with other packages’ way of
generating results, it is sometimes best to use it for individual chunks
only.
For data frames containing numerous variables, we can use the
max.tbl.height argument to wrap the results in a scrollable window
having the specified height, in pixels. For instance:
print(dfSummary(tobacco, valid.col = FALSE, graph.magnif = 0.75),
max.tbl.height = 300, method = "render")We can use the file argument with print() or view() to indicate
that we want to save the results in a file, be it html, Rmd, md,
or just plain text (txt). The file extension indicates to
summarytools what type of file should be generated.
view(iris_stats_by_species, file = "~/iris_stats_by_species.html")The append argument allows adding content to existing files generated
by summarytools. This is useful if you want to include several
statistical tables in a single file. It is a quick alternative to
creating an .Rmd document.
The following options can be set with
st_options():
| Option name | Default | Note |
|---|---|---|
| style | “simple” | Set to “rmarkdown” in .Rmd documents |
| plain.ascii | TRUE | Set to FALSE in .Rmd documents |
| round.digits | 2 | Number of decimals to show |
| headings | TRUE | Formerly “omit.headings” |
| footnote | “default” | Personalize, or set to NA to omit |
| display.labels | TRUE | Show variable / data frame labels in headings |
| bootstrap.css (*) | TRUE | Include Bootstrap 4 css in html outputs |
| custom.css | NA | Path to your own css file |
| escape.pipe | FALSE | Useful for some Pandoc conversions |
| subtitle.emphasis | TRUE | Controls headings formatting |
| lang | “en” | Language (always 2-letter, lowercase) |
(*) Set to FALSE in Shiny apps
| Option name | Default | Note |
|---|---|---|
| freq.totals | TRUE | Display totals row in freq() |
| freq.report.nas | TRUE | Display row and “valid” columns |
| ctable.prop | “r” | Display row proportions |
| ctable.totals | TRUE | Show marginal totals |
| descr.stats | “all” | “fivenum”, “common” or vector of stats |
| descr.transpose | FALSE | |
| descr.silent | FALSE | Hide console messages |
| dfSummary.varnumbers | TRUE | Show variable numbers in 1st col. |
| dfSummary.labels.col | TRUE | Show variable labels when present |
| dfSummary.graph.col | TRUE | Show graphs |
| dfSummary.valid.col | TRUE | Include the Valid column in the output |
| dfSummary.na.col | TRUE | Include the Missing column in the output |
| dfSummary.graph.magnif | 1 | Zoom factor for bar plots and histograms |
| dfSummary.silent | FALSE | Hide console messages |
| tmp.img.dir | NA | Directory to store temporary images |
st_options() # display all global options values
st_options('round.digits') # display the value of a specific option
st_options(style = 'rmarkdown') # change one or several options' values
st_options(footnote = NA) # Turn off the footnote on all outputs.
# This option was used prior to generating
# the present document.When a summarytools object is created, its formatting attributes are
stored within it. However, you can override most of them when using the
print() method or the view() function.
| Argument | freq | ctable | descr | dfSummary |
|---|---|---|---|---|
| style | x | x | x | x |
| round.digits | x | x | x | |
| plain.ascii | x | x | x | x |
| justify | x | x | x | x |
| headings | x | x | x | x |
| display.labels | x | x | x | x |
| varnumbers | x | |||
| labels.col | x | |||
| graph.col | x | |||
| valid.col | x | |||
| na.col | x | |||
| col.widths | x | |||
| totals | x | x | ||
| report.nas | x | |||
| display.type | x | |||
| missing | x | |||
| split.tables | x | x | x | x |
| caption | x | x | x | x |
| Argument | freq | ctable | descr | dfSummary |
|---|---|---|---|---|
| Data.frame | x | x | x | x |
| Data.frame.label | x | x | x | x |
| Variable | x | x | x | |
| Variable.label | x | x | x | |
| Group | x | x | x | x |
| date | x | x | x | x |
| Weights | x | x | ||
| Data.type | x | |||
| Row.variable | x | |||
| Col.variable | x |
Here’s an example in which we override 3 function-specific arguments, and one element of the heading:
(age_stats <- freq(tobacco$age.gr)) tobacco$age.gr
Type: Factor
| Freq | % Valid | % Valid Cum. | % Total | % Total Cum. | |
|---|---|---|---|---|---|
| 18-34 | 258 | 26.46 | 26.46 | 25.80 | 25.80 |
| 35-50 | 241 | 24.72 | 51.18 | 24.10 | 49.90 |
| 51-70 | 317 | 32.51 | 83.69 | 31.70 | 81.60 |
| 71 + | 159 | 16.31 | 100.00 | 15.90 | 97.50 |
| <NA> | 25 | 2.50 | 100.00 | ||
| Total | 1000 | 100.00 | 100.00 | 100.00 | 100.00 |
print(age_stats, report.nas = FALSE, totals = FALSE, display.type = FALSE,
Variable.label = "Age Group")tobacco$age.gr
Label: Age Group
| Freq | % | % Cum. | |
|---|---|---|---|
| 18-34 | 258 | 26.46 | 26.46 |
| 35-50 | 241 | 24.72 | 51.18 |
| 51-70 | 317 | 32.51 | 83.69 |
| 71 + | 159 | 16.31 | 100.00 |
Note that the original attributes are still part of the age_stats object, left unchanged.
- Options overridden explicitly in
print()orview()have precedence - Options specified as explicit arguments to
freq() / ctable() / descr() / dfSummary()come second - Global options set with
st_optionscome third
summarytools uses RStudio’s htmltools package and version 4 of Bootstrap’s cascading stylesheets.
It is possible to include your own css if you wish to customize the
look of the output tables. See ?print.summarytools for all the
details, but here is a quick example.
Say you need to make the font size really really small. For this, you would create a .css file - let’s call it “custom.css” - containing a class definition such as the following:
.tiny-text {
font-size: 8px;
}Then, to apply it to a summarytools object and display it in your browser:
view(dfSummary(tobacco), custom.css = 'path/to/custom.css',
table.classes = 'tiny-text')To display a smaller table that is not that small, you can use the
provided css class st-small.
To include summarytools functions in Shiny apps, it is recommended that you:
- set
bootstrap.css = FALSEto avoid interacting with the app’s layout - omit headings by setting the global option
headings = FALSE - adjust the size of the graphs in
dfSummary()using thedfSummary.graph.magnifglobal option - if
dfSummary()outputs are too wide, try omitting a column or two (valid.colandvarnumbers, for instance) - if needed, set the column widths manually with the
col.widthsparameter of theprint()method or theview()function
print(dfSummary(somedata, graph.magnif = 0.8),
method = 'render',
headings = FALSE,
bootstrap.css = FALSE)When generating markdown (as opposed to html) summaries in an .Rmd document, three elements are needed to display proper png graphs:
1 - plain.ascii is FALSE
2 - style is “grid”
3 - tmp.img.dir is defined
Why the third element? Although R makes it really easy to create temporary files and directories, they do have long pathnames, especially on Windows. Combine this with the fact that Pandoc currently determines the final (rendered) column widths by counting characters, including those of pathnames pointing to images. What we get is… some issues of proportion (!).
At this time, there seems to be only one solution around this problem: cut down on characters in pathnames. So instead of this:
+-----------+-------------------------------------------------------------------------+---------+
| Variable | Graph | Valid |
+===========+=========================================================================+=========+
| gender\ |  | 978\ |
| [factor] | | (97.8%) |
+----+---------------+----------------------------------------------------------------+---------+
…we aim for this:
+---------------+----------------------+---------+
| Variable | Graph | Valid |
+===============+======================+=========+
| gender\ |  | 978\ |
| [factor] | | (97.8%) |
+---------------+----------------------+---------+
Now CRAN policies are really strict when it comes to writing content in the user directories, or anywhere outside R’s temporary zone (for good reasons). So we need to let the users set this location themselves, therefore implicitly consenting to content being written outside R’s temporary zone.
On Mac OS and Linux, using “/tmp” makes a lot of sense: it’s short, and it’s self-cleaning. On Windows, there is no such convenient directory, so we need to pick one – be it absolute (“/tmp”) or relative (“img”, or simply “.”). Two things are to be kept in mind: it needs to be short (5 characters max) and we need to clean it up manually.
It is now possible to select the language used in the outputs. The following languages are available: English (en - default), French (fr), Spanish (es), Portuguese (pt), Turkish (tr), and Russian (ru). With the R community’s involvement, I believe we can add several more as time goes on.
To switch languages, simply use
st_options(lang = "fr")Any function will now produce outputs using that language:
view(freq(iris$Species))The language used for producing the object is stored within it as an attribute. This is to avoid problems when switching languages between the moment the object is stored, and the moment at which it is printed.
On most Windows systems, it will be necessary to change the LC_CTYPE
element of the locale settings if the character set is not included in
the current locale. For instance, in order to get good results – or
rather, any results at all – with the Russian language in a “latin1”
environment, we’ll need to do this:
Sys.setlocale("LC_CTYPE", "russian")
st_options(lang = 'ru')Then, to go back to default settings:
Sys.setlocale("LC_CTYPE", "")
st_options(lang = "en")Using the function use_custom_lang(), it is possible to add your own
set of translations. To achieve this, simply download the template csv
file from this
page,
customize the +/- 70 items, and call use_custom_lang(), giving it as
sole argument the path to the csv file you’ve created. Note that such
custom translations will not persist across R sessions. This means that
you should always have this csv file handy.
Sometimes, all you might want to do is change just a few keywords – say
you would rather have “N” instead of “Freq” in the title row of freq()
tables. No need to create a full custom language for that. Rather, use
define_keywords(). Calling this function without any arguments will
bring up, on systems that support graphical devices (the vast majority,
that is), an editable window allowing the modify only the desired items.
After closing the edit window, you will be offered to export the
resulting “custom language” into a .csv file that can be imported
later on with use_custom_lang().
Note that it is also possible to define one or several keywords using
arguments. For the list of all possible keywords to define, see
?define_keywords. For instance:
define_keywords(freq = "N")As stated earlier, version 0.9 brought many improvements to summarytools. Here are the key elements:
- Translations
- Improved printing of list objects
- Objects of class “stby” are automatically printed in the console
with optimal results; no more need for
view(x, method = "pander"); simply usestby()instead ofby() - Regular lists containing summarytools objects can also be
printed with optimal results simply by calling
print(x)(as opposed to “stby” objects, their automatic printing will not be optimal; that being said,freq()now accepts data frames as its first argument, so the need forlapply()is greatly reduced)
- Objects of class “stby” are automatically printed in the console
with optimal results; no more need for
- Easier management of global settings with
st_options()st_options()now has as many parameters as there are options to set, making it possible to set all options with only one function call; legacy way of setting options is still supported- Several global options were added, with a focus on simplifying Rmarkdown document creation
- Changes to
freq()- As mentioned earlier, the function now accepts data frames as
its main argument; this makes practically obsolete the use of
lapply()with it
- As mentioned earlier, the function now accepts data frames as
its main argument; this makes practically obsolete the use of
- Improved outputs when using
stby() - Changes to
ctable()- Fully supports
stby() - Improved number alignment
- Added “chisq” parameter
- Fully supports
- Changes to
descr()- For the
statsargument, Values “fivenum” and “common” are now allowed, the latter representing the collection of mean, sd, min, med, max, n.valid, and pct.valid - Improved outputs when using
stby() - The variable used for weights (if any) is removed automatically from the data so no stats are produced for it
- For the
- Changes to
dfSummary()- Now fully compatible with Rmarkdown
- Number of columns is now included in the heading section
- Number of duplicated rows is also shown in the heading section
- Bar plots now more accurately reflect counts, as they are not stretched across table cells (this allows the comparison of frequencies across variables)
- Columns with particular content (unary/binary, integer sequences, UPC/EAN codes) are treated differently; more relevant information is displayed, while irrelevant information is hidden
- For html outputs, a new parameter
col.widthscan be used to set the width of the resulting table’s columns; this addresses an issue with some graphs not being shown at the desired magnification level (although much effort has been put into improving this as well) max.tbl.heightparameter added
For a preview of what’s coming in the next release, see the development branch.
The package comes with no guarantees. It is a work in progress and feedback / feature requests are welcome. Just send me an email (dominic.comtois (at) gmail.com), or open an issue if you find a bug or wish to submit a feature request.
Also, the package grew significantly larger, and maintaining it all by myself is time consuming. If you would like to contribute, please get in touch, I’d greatly appreciate the help.